MEC_4891_sm_FigS1-5andTableS1
Supplementary Information to the manuscript entitled:
Ecometagenetics Confirms High Tropical Rainforest Nematode Diversity
DL Porazinska, RM Giblin-Davis, A Esquivel, TO Powers, W Sung & WK Thomas
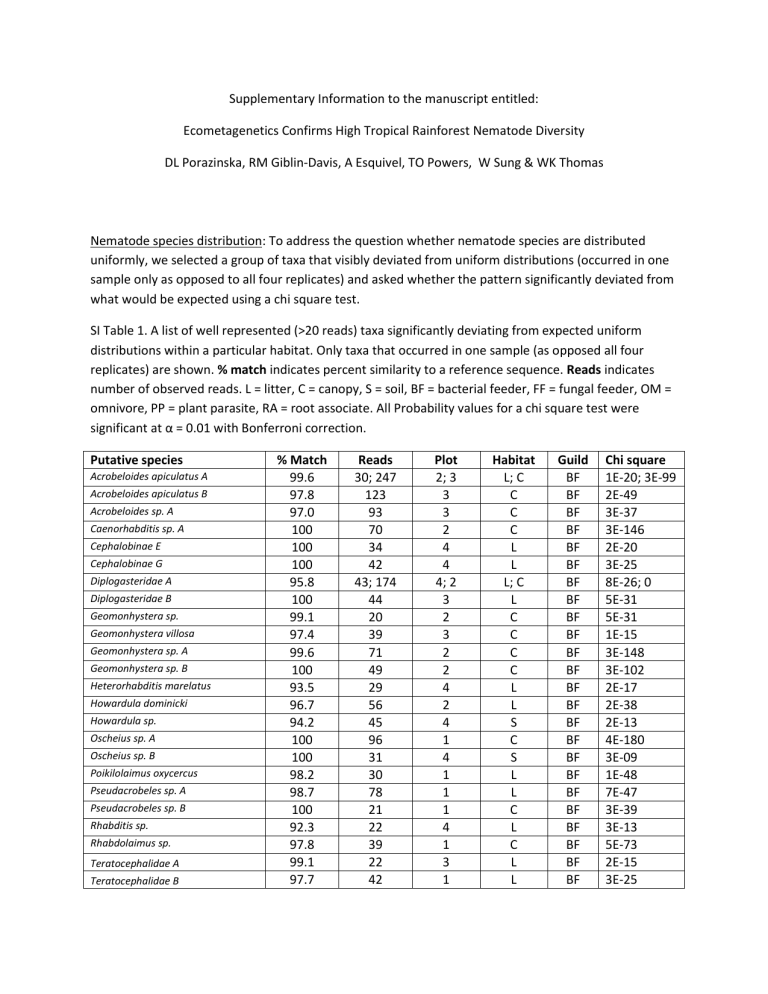
Nematode species distribution: To address the question whether nematode species are distributed uniformly, we selected a group of taxa that visibly deviated from uniform distributions (occurred in one sample only as opposed to all four replicates) and asked whether the pattern significantly deviated from what would be expected using a chi square test.
Putative species
Acrobeloides apiculatus A
Acrobeloides apiculatus B
Acrobeloides sp. A
Caenorhabditis sp. A
Cephalobinae E
Cephalobinae G
Diplogasteridae A
Diplogasteridae B
Geomonhystera sp.
Geomonhystera villosa
Geomonhystera sp. A
Geomonhystera sp. B
Heterorhabditis marelatus
Howardula dominicki
Howardula sp.
Oscheius sp. A
Oscheius sp. B
Poikilolaimus oxycercus
Pseudacrobeles sp. A
Pseudacrobeles sp. B
Rhabditis sp.
Rhabdolaimus sp.
Teratocephalidae A
Teratocephalidae B
SI Table 1. A list of well represented (>20 reads) taxa significantly deviating from expected uniform distributions within a particular habitat. Only taxa that occurred in one sample (as opposed all four replicates) are shown. % match indicates percent similarity to a reference sequence. Reads indicates number of observed reads. L = litter, C = canopy, S = soil, BF = bacterial feeder, FF = fungal feeder, OM = omnivore, PP = plant parasite, RA = root associate. All Probability values for a chi square test were significant at α = 0.01 with Bonferroni correction.
98.7
100
92.3
97.8
99.1
97.7
99.1
97.4
99.6
100
93.5
96.7
94.2
100
100
98.2
% Match
99.6
97.8
97.0
100
100
100
95.8
100
78
21
22
39
22
42
29
56
45
96
31
30
20
39
71
49
Reads
30; 247
123
93
70
34
42
43; 174
44
1
1
4
1
3
1
4
2
4
1
4
1
2
3
2
2
Plot
2; 3
3
3
2
4
4
4; 2
3
L
C
L
C
L
L
S
C
S
L
L
L
C
C
C
C
Habitat Guild Chi square
L; C BF 1E-20; 3E-99
C
C
BF
BF
2E-49
3E-37
C
L
L
L; C
L
BF
BF
BF
3E-146
2E-20
3E-25
BF 8E-26; 0
BF 5E-31
BF
BF
BF
BF
BF
BF
BF
BF
BF
BF
BF
BF
BF
BF
BF
BF
5E-31
1E-15
3E-148
3E-102
2E-17
2E-38
2E-13
4E-180
3E-09
1E-48
7E-47
3E-39
3E-13
5E-73
2E-15
3E-25
Teratocephalidae C
Teratocephalus lirellus
Tripylina sp.
Trischistoma monohystera
Aphelenchoididae
Schistonchus guangzhouensis
Telotylenchinae
Tylencholaimus terestris
Tylencholaimus D
Chrysonema sp. A
Dorylaimellus sp.
Drepanodorylaimus flexus
Labronemella sp.
Laimydorus sp.
Mesodorylaimus pseudobastiani
Paractinolaiminae A
Paractinolaimus sp.
Qudsianematinae B
Anguininae
Anguininae A
Anguininae B
Anguininae C
Ditylenchus destructor
Ditylenchus sp.
Nothocriconemoides sp.
Xiphinema brasiliense
Filenchus sp.
Tylenchinae A
92.4
98.7
100
98.7
100
92.9
100
97.7
100
100
100
97.5
94.4
92.0
99.6
100
99.6
100
100
92.3
100
94.25
97.0
95.6
100
100
92.7
100
37
22
33
192
23
23
32
208
28
22
49
113
23
49
49
721
43
512
32
48
147
34
28; 26
157
86
90
74
32
4
1
3
3
3
2
4
2
3
1
1
2
4
4
3
4
4
4
3
3
3
3
1; 4
4
1
1
1
4
L
L
S
C
L
C
S
L
L
C
C
C
C
C
C
C
C
C
L
C
L
C
L; C
C
S
C
L
L
Linking OCTUs to species: the details for recognizing OCTU-species relationships are presented in
Porazinska et al 2010b. Using data from control experiments, we determined that each species, whether represented by 1or >1 individuals is illustrated by a series of sequences that could be clustered to 2 main groups: Head and Tail. Head is a cluster of most abundant and dominant type of sequences and Tail is a series of clusters of rare sequences that are variant to the dominant type sequence. In order to understand OCTU-species-abundance relationships, all sequences within the Head and Tail clusters
(OCTUs) need to be summed up. Figure 1 illustrates how this is being accomplished.
Figure 1. An example of linking OCTUs (clusters of sequences grouped at 99% sequence similarity) to species and abundance data using a single putative taxon, i.e., all 18 OCTUs give a best match to an
Anguinid D species. OCTU 313 is the Head OCTU with a total of 251 sequencing reads, length of 220 bp, and matching our consensus reference sequence at a BitScore of 436 at 100% along the entire length of
BF 3E-22
BF 3E-13
BF 7E-27
BF 0
FF 3E-16
FF 7E-48
FF 1E-09
FF 1E-143
FF 9E-20
OM 4E-41
OM 9E-22
OM 3E-236
OM 4E-06
OM 9E-13
OM 1E-19
OM 3E-188
OM 3E-11
OM 1E-133
PP 1E-22
PP 3E-19
PP 5E-104
PP 1E-13
PP 8E-17; 7E-07
PP 8E-41
PP 7E-74
PP 7E-169
RA 2E-44
RA 3E-19
the OCTU. Tail OCTUs (e.g. OCTU 10) with considerably fewer than Head OCTU sequencing reads match the same reference sequence but at slightly lower BitScore and % similarity values. All 18 Head and Tail
OCTUs are linked to 1 (Anguinid D) species. To reflect the abundance relationships among samples, all reads of the Head and Tail OCTUs are summed up to give an overall number of reads per species per sample. S = soil, L = litter, C = canopy. 1-4 indicate the number of the sampling plot.
14490
15665
16319
11733
10801
13654
17632
7427
OCTU Sequences OCTU Length BitScore E-Value MatchBases TotalBases PercentID DBmatch S1 S2 S3 S4 L1 L2 L3 L4 C1 C2 C3 C4
313 251 220 436 e-119 220 220 100 Anguininae_D_Juv_804L-005 0 0 41 44 39 29 15 39 31 13
7956
10175
189
10
31
2
216
216
227
414
410
404 e-113 e-111 e-110
213
214
223
214
216
229
99.53
Anguininae_D_Juv_804L-005 0 0 0 2 0 1 0 3 2 2
99.07
Anguininae_D_Juv_804L-005 0 0 5 5 4 0 4 9 3 1
97.38
Anguininae_D_Juv_804L-005 0 0 1 0 1 0 0 0 0 0
9602
12178
1273
7827
15615
16110
1
3
1
1
2
2
220
219
214
211
214
214
402
400
398
390
390
381 e-109 e-108 e-108 e-105 e-105 e-103
217
216
211
206
211
209
221
220
214
209
215
214
98.19
Anguininae_D_Juv_804L-005 0 0 1 0 0 0 0 0 1 0
98.18
Anguininae_D_Juv_804L-005 0 0 0 0 1 1 0 0 0 0
98.6
Anguininae_D_Juv_804L-005 0 0 1 0 0 0 0 0 0 0
98.56
Anguininae_D_Juv_804L-005 0 0 1 0 1 1 0 0 0 0
98.14
Anguininae_D_Juv_804L-005 0 0 0 0 0 1 0 0 0 0
97.66
Anguininae_D_Juv_804L-005 0 0 0 1 0 0 0 0 0 0
1
1
1
1
1
1
1
2
213
200
205
210
210
202
206
218
369
361
353
343
341
299
4.00E-99
8.00E-97
2.00E-94
349 3.00E-93
345 5.00E-92
2.00E-91
8.00E-91
3.00E-78
207
196
199
195
196
195
195
194
213
200
205
201
203
202
202
208
97.18
98
97.07
96.53
96.53
93.27
Anguininae_D_Juv_804L-005
Anguininae_D_Juv_804L-005
Anguininae_D_Juv_804L-005
97.01
Anguininae_D_Juv_804L-005 0 0 0 0 0 1 0 0 0 0
96.55
Anguininae_D_Juv_804L-005 0 0 1 0 0 0 0 0 0 0
Anguininae_D_Juv_804L-005
Anguininae_D_Juv_804L-005
Anguininae_D_Juv_804L-005
0
0
0
0
0
0
0
0
0
0
0
0
0
0
1
1
1
0
1
0
0
0
0
0
0
0
0
0
0
1
0
0
0
0
0
0
0
0
0
0
0
1
0
0
0
0
0
0
0
1
0
0
0
0
0
0
0
0
0
0
Sum of Head and Tail reads within each sample
S1 S2 S3 S4 L1 L2 L3 L4 C1 C2 C3 C4
0 0 0 0 54 53 47 34 20 51 38 16
Head OCTU
Tail OCTUs
Read abundance vs. nematode abundance: Normalization of the read abundance data was necessary for derivation of ACE and ICE diversity indices that are very sensitive to the presence of unusually high variation in the read abundance data (e.g. 5 vs. 15000 reads). Although the relationship between the number of sequencing reads and the number of nematode individuals is rather complex (e.g. it appears species specific and dependent on the rDNA copy numbers), we used general guidelines provided by our control experiments (Porazinska et al 2009, 2010a) to bring raw read data into the context of more meaningful variation than we typically observe in the nematode abundance data. Read data and equivalent numbers of individuals are provided within the figure.
Figure 2. Transformation of the read abundance data to the number of nematode individuals.
300
200
100
0
700
600
500
400
18000
16000
14000
12000
10000
8000
6000
4000
2000
0
Reads
6 - 400
401 - 750
751 - 1000
1001 - 2000
2001 - 2500
2501 - 4000
4001 - 10000
10001 - 12000
Nematodes
8
10
15
33
1
2
3
40
120
100
80
60
40
20
0
S1 S2 S3 S4 L1 L2 L3 L4 C1 C2 C3 C4
Certainty of taxonomic identity: To illustrate the strength of the taxonomic identity of OCTUs, we clustered all OCTUs into categories representing % identity match (100% -90%) to a sequence in the database for all OCTUs (Heads and Tails) and Head OCTUs only. In both cases, the great majority of
OCTUs gave a match at >95% strongly indicating taxonomic membership.
Figure 3. Distribution of nematode OCTUs (Total vs. Head) along the % identity match to a sequence in the reference database.
100%
99%
98%
97%
96%
95%
94%
93%
92%
91%
90%
Total Head
Certainty of taxonomic assignments: We placed all putative nematode species into their respective feeding guilds. Because the majority of these species (140) were not identified by a perfect (100%) match at the GenBank database, we wondered about the quality of these assignments. In order to resolve this uncertainty, we checked the assignments in two ways:
1. Among all putative nematode species (214), there was a subset of 74 species that gave a 100% match to our internal database generated during our earlier studies using traditional identification protocols and whose identity and guild assignment therefore was known. Because these 74 species represented a random subset of a larger species pool, we hypothesized that the subset of the 74 species should be illustrative of the larger species pool. Figure 1 illustrates this approach.
2. We used the sequences of the 74 species and blast-matched them to the GenBank sequences to verify their taxonomic assignments in the hypothetical absence of an internal database. Although the % identity decreased slightly below 100%, the taxa still fell into the same feeding guild categories (data not shown) indicating robustness of the assignments.
Figure 4. Distribution of nematode species within each trophic group for total observed number of species (Total) and a subset of species that 100% matched our internal database from Costa Rica (100%
Match). BF = bacterial feeders, FF = fungal feeders , OM = omnivorous nematodes, PP = plant parasites,
PR = predators, RA = root associates.
120
100
80
60
BB
FF
OM
PP
PR
RA
40
20
0
Total 100% Match
Nematode species distributions: To determine how cosmopolitan vs. rare nematode species are, we looked at the presence/absence of each putative species across all 12 samples (4 soil, 4 litter, and 4 canopy). Cosmopolitan species would tend to occur across majority of the samples, and rare species would tend to occur across very few samples.
Figure 5. Occurrence of a nematode species across all 12 samples.
100
90
80
70
60
50
40
30
20
10
0
1 2 3 4 5
Number of Samples
6 7 8 9