Supplementary Data
advertisement

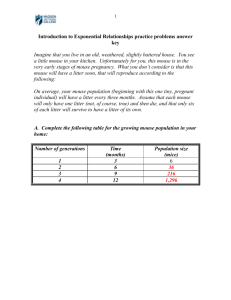
Supplementary Materials and Methods The present study was conducted in accordance with the Guiding Principles for the Care and Use of Laboratory Animals, as adopted by the Committee on Animal Research of Hoshi University, which is accredited by the Ministry of Education, Culture, Sports, Science and Technology of Japan. Every effort was made to minimize the numbers and any suffering of animals used in the following experiments. Animals were used only once in the present study. All behavioral experiments were conducted in a single-blind fashion to avoid the effect of subjectivity. Animals All animals were housed at a room temperature of 22 ± 1˚C with a 12-h light-dark cycle (light on 8:00 am to 8:00 pm). Food and water were available ad libitum. Intrathecal injection Intrathecal (i.t.) injection was performed as described by Hylden and Wilcox (1980)1 using a 25-l Hamilton syringe (Hamilton Company, NV, USA) with a 30 1/2-gauge needle. The needle was inserted into the intervertebral space of the spinal cord of unanesthetized mice. A reflexive flick of the tail was considered to indicate the accuracy of each injection. The volume for intrathecal injection was 4 l. Neuropathic pain model Mice were anesthetized with 3% isoflurane. We produced a partial sciatic nerve ligation model by tying a tight ligature with 8-0 silk suture around approximately 1/3 to 1/2 the diameter of the sciatic nerve on the right side (ipsilateral side) of mice under a light microscope (SD30, Olympus, Tokyo, Japan), as described previously2. In sham-operated mice, the nerve was exposed without ligation. Gene transcription profiling using the spinal cord of nerve-ligated and sham-operated mice In the present study, samples were prepared 7 days after partial sciatic nerve ligation or sham operation as a control. For sample preparation, the ipsilateral side of the mouse spinal cord was quickly removed after decapitation and rapidly transferred to a tube filled with RNA stabilization reagent (RNAlater; QIAGEN, Heiden, Germany). Total RNA of the spinal cord was prepared using an RNeasy Lipid Tissue Mini Kit (QIAGEN) according to the manufacturer’s instructions. The RNA concentration was measured at 260 nm with a NanoDrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies Inc., Rockland, DE, USA). RNA was subjected to quality analysis on a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) using an RNA 6000 NanoChip. For microarray-based expression analysis, cRNA labeled with cyanine 3 was synthesized from 500 ng of total RNA using a low RNA input fluorescent linear amplification kit (Agilent Technologies). cRNA quantity and quality were measured using a NanoDrop and 2100 Bioanalyzer. Aliquots of 1.65 g of fragmented cRNA were hybridized to G4122F whole mouse genome microarrays (Agilent Technologies) following the standard protocol (version 4.0) provided by the manufacturer. The slides were scanned with a DNA microarray scanner (Agilent Technologies). Scanned images were analyzed with Feature Extraction software (Agilent Technologies). The obtained expression data were imported into GeneSpring GX 7.0 software (Agilent Technologies) for normalization and data analysis. For per chip normalization, the 50 th percentile of the signal values taken from each microarray was used as the normalizing reference. Per gene normalization was based on calculating the fold expression levels relative to the median in sham-operated mice (n = 3). The fold change in gene expression was calculated as follows: sciatic nerve ligation/sham operation. Tissue dissection and preparation of the cytosol fraction Tissue dissection and preparation of the cytosol fraction were performed following the methods described previously3. 1, 3, 5, 7, 14 and 28 days after the surgery, the lumbar spinal cord after nerve ligation or sham operation was quickly removed by decapitation and dissected on an ice-cold metal plate. The tissue was homogenized in 10 volumes of ice-cold buffer containing 20 mM Tris–HCl (pH 7.5), 2 mM ethylene diamine tetraacetic acid (EDTA), 0.5 mM ethylene glycol tetraacetic acid (EGTA), 1 mM phenylmethylsulfonyl fluoride, 25 g of leupeptin per ml, and 10 g of aprotinin per mL, 10 mM Sodium fluoride, 1 mM sodium orthovanadate, and 0.32 M sucrose. The homogenate was centrifuged at 1000 x g for 10 min at 4 °C and the supernatant was ultracentrifuged at 100,000 x g for 30 min at 4°C. The resulting supernatant was retained as the cytosolic fraction. Western blotting The protein levels were performed following the methods described previously3,4. An aliquot of tissue sample was diluted with an equal volume of 2 x electrophoresis sample buffer (EZ Apply, Atto Co., Tokyo, Japan). Proteins were separated by size on 5-20% sodium dodecyl sulfate (SDS)-polyacrylamide gradient gel or 15% tris SDS-tricine polyacrylamide homogenous gel using the buffer system (10 x TG-SDS READY PACK, Amersco, Olt, USA). After transferring onto nitrocellulose membranes, membranes were blocked in Tris-buffered saline (TBS) containing 0.3-1.0% nonfat milk (Bio-Rad Laboratories, Hercules, CA, USA) containing 0.1% Tween 20 (Research Biochemicals, Inc., MA, USA), for 1 h at room temperature with agitation and 4 h at 4°C with stationary. The membrane was incubated with the following primary antibodies; anti-signal tranducer and activator of transcription 3 (STAT3) (rabbit polyclonal, 1:1000, Cell signaling Technology, Inc., MA, USA), anti-phosphorylated-STAT3 (rabbit polyclonal, 1:1000, Cell signaling Technology, Inc., MA, USA), anti-ionized calcium binding adaptor molecule 1 (Iba-1) (rabbit polyclonal 1:1000, Wako pure Chemicals, industrie, Ltd, Osaka, Japan), anti-phosphorylated-extracellular signal-regulated kinase (p-ERK) (mouse monoclonal, 1:500, Santa cruz biotechnology, inc., CA, USA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (mouse monoclonal, 1:400,000, Chemicon International, Inc.) overnight at 4°C. The membrane was washed in TBS containing 0.05% Tween 20 (TTBS), followed by 2 h of incubation at room temperature with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG or HRP-conjugated anti-mouse IgG (Southern Biotechnology Associates, Inc., Birmingham, AL, USA) diluted 1:10,000 in TBS containing 0.3-1.0% nonfat milk containing 0.1% Tween 20. After this incubation, the membranes were washed in TTBS. The antigen–antibody peroxidase complex was then finally detected by enhanced chemiluminescence (Pierce, Rockford, IL, USA) according to the manufacturer’s instructions and visualized by exposure to Amersham Hyperfilm (Amersham Life Sciences, Arlington Heights, IL, USA). RNA preparation and semi-quantitative analysis by reverse transcription-polymerase chain reaction Total RNA obtained from the mouse spinal cord was extracted using the SV Total RNA Isolation system (Promega Co., Madison, WI, USA) as described previously 5. After animals were decapitated, L3-5 lumbar dorsal root ganglia (DRGs) were rapidly removed, and then the attached sciatic nerve and dorsal roots were transected adjacent to the ganglion. Total RNA obtained from the mouse DRG was extracted using ISOGENE (NIPPON GENE Co., Tokyo, Japan) according to the manufacturer’s instructions. First- of a PCR solution containing 0.8 mM MgCl , dNTP mix, and DNA polymerase with synthesized primers of MCP-3 (sense: 5'-TCT GTG CCT GCT GCT CAT AG-3', antisense: 5'-CTT TGG AGT TGG GGT TTT CA-3'), MCP-1 (sense: 5'-AGC CAG ATG CAG TTA ACG C-3', antisense: 5'-CTG ATC TCA TTT GGT TCC GA-3'), RANTES (sense: 5’-GGT ACC ATG AAG ATC TCT GCA-3’, antisense: 5’-AAA CCC TCT ATC CTA GCT CAT-3’), PU.1 (sense: 5’-CGG ATG TGC TTC CCT TAT CAA AC-3’, antisense: 5’-TGA CTT TCT TCA CCT CGC CTG TC-3’), NF-IL-6 (sense: 5’-GGC CCT GAG TAA TCA CTT AAA GA-3’, antisense: 5’-ACA TAC GCC TCT TTT CTC ATA G-3’), IL-6 (sense: 5'-ATG AAG TTC CTC TCT GCA AGA GAC T-3', antisense: 5'-CAC TAG GTT TGC CGA GTA GAT CTC-3'), leukemia inhibitory factor (LIF) (sense: 5'-GGC AAC CTC ATG AAC CAG ATC A-3', antisense: 5'-GCA AAG CAC ATT GCT GAG GAG GC-3'), ciliary neurotrophic factor (CNTF) (sense: 5'-TGG CTA GCA AGG AAG ATT CG-3', antisense: 5'-ATT GCC TGA TGG AAG TCA CC-3'), cardiotrophin-1 (CT-1) (sense: 5'-CGG CCA ACA GCA CTG CAG GCA TC-3', antisense: 5'-AAG CTC CCT GCA GAG AGG AGA GC-3'), oncostatin-M (OSM) (sense: 5'-AAC ACT GCT CAG TTT GAC CC-3', antisense: 5'-ACA GTG CTC AGG AAG TGA GG-3') or GAPDH (sense: 5’-CCC ACG GCA AGT TCA ACG G-3’, antisense: 5’-CTT TCC AGA GGG GCC ATC CA-3’). Samples were heated to 94˚C for 3 min, 55˚C for 1 min, and 72˚C for 1 min, and then subjected to 29 cycles of 94˚C for 30 sec, 55˚C for 1 min, and 72˚C for 1 min. The final incubation was at 72˚C for 7 min. The mixture was subjected to 2% agarose gel for electrophoresis with the indicated markers and primers for the internal standard (GAPDH). Each sample was applied to more than two lanes in the same gel. The agarose gel was stained with ethidium bromide and photographed with UV transillumination. The intensity of the bands was analyzed and quantified by computer-assisted densitometry using the image-analysis software ImageJ. For the control, the different intensities of each band from the sample of sham-operated mice or sham-operated wild-type mice were analyzed, and the average intensity was calculated. Next, each control intensity was again compared with the average intensity to calculate the standard error. Under these conditions, the intensities of bands for samples obtained from nerve-ligated mice were analyzed and compared with the average intensity for sham-operated mice. Finally, the % of the control with the standard error for each sample was quantified. Quantitative analysis by Real-time reverse transcription-polymerase chain reaction A KAPA SYBR® FAST qPCR kit (Kapa Biosystems, Inc., MA, USA) was used as the basis for the reaction mixture in the real-time PCR assay. Each gene prepared by the above procedure was amplified in 20 l of a PCR solution containing 10 l of the SYBR® FAST qPCR Master Mix with synthesized primers for MCP-3 (sense: 5'-TCT GTG CCT GCT GCT CAT AG-3', antisense: 5'-CTT TGG AGT TGG GGT TTT CA-3') or -actin (sense: 5’-CAGCTTCTTTGCAGCTCCTT-3’, antisense: 5’-TCACCCACATAGGAGTCCTT-3’). In addition to each sample, each test run included a no-target control that contained reaction mixture and PCR-grade water. RT-PCR with a Mastercycler EP realplex (Eppendorf, Wesseling Berzdorf, Germany) was performed with the following cycling conditions: 94˚C for 3 min, 55˚C for 1 min, and 72˚C for 1 min, followed by cycled 29 cycles of 94˚C for 30 sec, 55˚C for 1 min, and 72˚C for 1 min. The final incubation was at 72˚C for 7 min. Fluorescence detection was conducted after each extension step. Chromatin immunoprecipitation A chromatin immunoprecipitation (ChIP) assay was performed as described previously 6,7 with minor modifications. Briefly, the ipsilateral side of the mouse spinal cord tissue was dissected as described above and cross-linked, and then tissue was lysed. Fifteen μg of sonicated chromatin extracted from the mouse spinal cord was diluted in 1 mL ChIP dilution buffer (1.1%w/v DOC, 1.1%w/v Triton X-100, 167 mM NaCl, 50 mM Tris-HCl pH8.0) and then immunoprecipitated using 2 μg of specific antibodies against acetylated histone H3 (Millipore), H3K4 trimethylation (Wako Pure Chemicals), H3K9 trimethylation (Millipore), and H3K27 trimethylation (Millipore), overnight at 4ºC. The immuno-complex was collected by Dynabeads Protein A (Invitrogen Dynal AS, Oslo, Norway), and DNA was recovered with RNaseA treatment, Proteinase K treatment followed by isopropanol precipitation. Immunoprecipitated DNA was dissolved in 50 l of 1 x TE and 1 l was used for quantitative PCR. Quantitative PCR was performed as described previously8. The primer sequences were; MCP-3 (sense: 5’-CCT GTT TCT AGA CTC CAA GCT CC-3’, antisense 5’-ATT CCA ACC AGC TCA GCT ACT AT-3’), MCP-1 (sense: 5’-ATT TGC TCC CAG GAG TGG C-3’, antisense 5’-GGA GTC AGG CAG GGT GCT T-3’), RANTES (sense: 5’-CTG GAC TGG AGG GCA GTT AG-3’, antisense 5’-TTG GGG AGT TTC CAC AAA AG-3’). Immunohistochemistry using the mouse spinal cord-slice sections One to 2 weeks after partial sciatic nerve ligation, mice were deeply anesthetized with 3% isoflurane and intracardially perfusion-fixed with freshly prepared 4% paraformaldehyde in 0.1 M in phosphate-buffered saline (PBS, pH 7.4). After perfusion, the sikn and viscera were removed. Then, the ribs were counted, and the most caudal rib marked with ink. The most craniad vertebra that lacked an articulation with a rib at its rostral margin was regarded as the L1 vertebra, and was also marked. Further dissection was performed to expose the vertebral and sacral bones in order to identify the sites of fusion of the lower lumbar vertebrae. Then, L4 spinal cord was quickly removed. The lumbar spinal cord sections were post-fixed in 4% paraformaldehyde for 2 h, and permeated with 20 (w/v)% sucrose solution in 0.1 M PBS for 24 h and 30 (w/v)% sucrose solution in 0.1 M PBS for 48 h at 4˚C with agitation. The lumbar spinal cord sections were then frozen in an embedding compound (Sakura Finetechnical, Tokyo, Japan) on isopentane using liquid nitrogen and stored at -30˚C until use. Frozen segments were cut with a freezing cryostat (Leica CM 1510, Leica Microsystems AG, Wetzlar, Germany) at a thickness of 10 m and thaw-mounted on poly-L-lysine-coated glass slides. The sections were blocked in 7% normal horse serum (NHS) or 10% normal goat serum (NGS) in 0.01 M PBS for 1 h at room temperature and then incubated for 48 h at 4˚C with the following primary antibodies: anti-MCP-3 (goat polyclonal, 1:200, R&D systems, Inc., MN, USA), anti-neuron-specific nuclear protein NeuN (mouse monoclonal, 1:250, Millipore Co., MA, USA), anti-glial fibrillary acidic protein (GFAP) (rabbit polyclonal, 1:40, Nichirei Bioscience Co, Tokyo, Japan), anti-Iba-1 (rabbit polyclonal, 1:200, Wako Pure Chemical, Industrie, Ltd., Japan), anti-metabotropic glutamate subtype 5 receptor (mGluR5) (rabbit polyclonal, 1:3000, Upstate USA, Inc., VA, USA), anti-protein kinase C (PKC) (rabbit polyclonal, 1:1000, Santa Cruz Biotechnology) and anti-microtubule associated protein-2 (MAP2) (mouse monoclonal, 1:300, Chemicon International, Inc.). The antibody was then rinsed and incubated with an appropriate secondary antibody conjugated with Alexa FluorTM 488 and/or 546 (Molecular Probes, Inc., Eugene, OR, USA) for 2 h at room temperature. The slides were then cover-slipped with Dako Huorescent mounting medium (Dako, Denmark). All sections were observed with a light microscope (Olympus BX-53; Olympus) and photographed with a digital camera (CoolSNAP HQ; Olympus). Digitized images of superficial laminae of the spinal dorsal horn sections were captured at a resolution of 1316 x 1035 pixels with camera. Preparation of spinal microglial cells Purified spinal glial cells were grown as follows. The mouse spinal cord was obtained from newborn ICR mice, minced, and treated with trypsin (0.025%, Invitrogen, Carlsbad, CA) dissolved in PBS. After enzyme treatment at 37˚C for 15 min, cells were dispersed by gentle agitation through a pipette and plated on a flask. Four weeks after culture, floating microglia was collected from cultures by shaking flasks, then the cells were seeded at a density of 5 x 104 cells/mL. The cells were maintained for 3 days in Dulbecco's Modified Eagle Medium (DMEM) (Life Technologies Japan Ltd., Tokyo, Japan) supplemented with 5% precolostrum newborn calf serum (Life Technologies Japan), 5% heat-inactivated (56 ˚C, 30 min) horse serum (Life Technologies Japan), 10 U/ml penicillin and 10 g/ml streptomycin in a humidified atmosphere of 95% air and 5% CO2 at 37˚C. These cells were treated with normal medium or recombinant MCP-3 (10 ng/mL) with or without a CCR2 antagonist RS102895 (10 M) for 1 day, and used as activated glial cells in the present study. Immunohistochemistry using purified spinal microglia Purified spinal microglia were identified by immunofluorescence using anti-Iba-1 (1:250, Wako Pure Chemicals) followed by incubation with an appropriate secondary antibody conjugated with Alexa 488 (Molecular Probes). Stained cells were mounted on glass slides and viewed using an Olympus BX-53 (Olympus). Each experimental condition was repeated for three independent culture preparations. Measurement of thermal hyperalgesia To assess the sensitivity to thermal stimulation, each of the hind paws of mice was tested individually using a thermal stimulus apparatus (model 33 Analgesia Meter; IITC Inc./Life Science Instruments, Woodland Hills, CA, USA). The intensity of the thermal stimulus was adjusted to achieve an average baseline paw-withdrawal latency of approximately 8 to 10 sec in naive mice. Only quick hind paw movements (with or without licking of the hind paws) away from the stimulus were considered to be a withdrawal response. Paw movements associated with locomotion or weight-shifting were not counted as a response. The paws were measured alternating between the left and right with an interval of more than 3 min between measurements. The latency of paw withdrawal after the thermal stimulus was determined as the average of 3 trials per paw on each side. Before the behavioral responses to the thermal stimulus were tested, mice were habituated for 1 h in an acrylic cylinder (15 cm high and 8 cm in diameter). Under these conditions, the latency of paw withdrawal in response to a thermal stimulus was tested. The latency of paw withdrawal in response to a thermal stimulus was measured before and after partial sciatic nerve ligation. To investigate the effect of a single intrathecal treatment with MCP-3 (0.1, 0.3, 1 pmol/mouse) and IL-6 (1 pmol/mouse) in normal mice or intrathecal treatment with anti-MCP-3 antibody, a selective CCR2 antagonist, RS102895 and recombinant mouse gp130/Fc chimera (gp130/Fc) in nerve-ligated mice on the thermal latency, the latency of paw withdrawal was measured before and after injection until the latency returned to the baseline. Measurement of tactile allodynia To quantify the sensitivity to a tactile stimulus, paw withdrawal in response to a tactile stimulus was measured using von Frey filaments (North Coast Medical, Inc., Morgan Hill, CA, USA) with a bending force of 0.02 g. The von Frey filament was applied to the plantar surface of the hind paw for 3 sec, and this was repeated 3 times at intervals of at least 5 sec. Each of the hind paws was tested individually. Paw withdrawal in response to a tactile stimulus was evaluated by scoring as follows: 0, no response; 1, a slow and slight withdrawal response; 2, a slow and prolonged flexion withdrawal response (sustained lifting of the paw) to the stimulus; 3, a quick withdrawal response away from the stimulus without flinching or licking; 4, an intense withdrawal response away from the stimulus with brisk flinching and/or licking. The latency of paw withdrawal in response to a tactile stimulus was determined as the average of 2 trials per paw on each side. Paw movements associated with locomotion or weight-shifting were not counted as a response. The paws were measured alternating between left and right with an interval of more than 3 min between measurements. Before the behavioral responses to a tactile stimulus were tested, mice were habituated for 1 h on an elevated nylon mesh floor. Under these conditions, paw withdrawal in response to a stimulus was measured before and after partial sciatic nerve ligation. To investigate the effect of a single intrathecal treatment with MCP-3 (0.1, 0.3, 1 pmol/mouse) and IL-6 (1 pmol/mouse) in normal mice or intrathecal treatment with anti-MCP-3 antibody, a selective CCR2 antagonist, RS102895 and recombinant mouse gp130/Fc chimera (gp130/Fc) in nerve-ligated mice on the tactile latency, the latency of paw withdrawal was measured before and after injection until the latency returned to the baseline. Functional imaging Experiments were performed with a Unity Inova spectrometer (Varian, Palo Alto, CA) which was interfaced to a 9.4-T/31-cm horizontal bore magnet equipped with actively shielded gradients capable of 300 mT/m in a risetime of 500 sec (Magnex Scientific, Abingdon, UK). During the measurements, mice were slightly anaesthetized with isoflurane (0.5 - 1%). Mice were then transferred to a cradle designed to fit inside the probe of the MR system. A continuous fMRI scanning protocol was used to study changes in brain signal intensity using T2-weighted blood oxygenation level-dependent (BOLD) contrast. A functional series was acquired using the Echo Planar Imaging Technique (EPI: matrix = 64 x 64, TR = 2000 ms, TE = 35 ms, 2 acquisitions, slice thickness = 1 mm, field of view = 25.6 x 25.6 mm2). Anatomical scans with a high spatial resolution were collected using a fast spin echo pulse sequence (matrix = 256 x 256, TR = 2000 ms, TE = 45 ms, slice thickness = 1mm, field of view = 25.6 x 25.6 mm2). To investigate the effect of a single intrathecal treatment with MCP-3 (1 pmol/mouse), ICR mice were anesthetized at 3 h after intrathecal injection of MCP-3, and heat stimulation was applied to the right hindpaw. Contact heat stimulation was performed using a custom-made, computer-controlled Peltier heating and cooling device. Peltier elements with a surface measuring 8.3 x 8.3 mm were fixed at the right hindpaw. Starting at a baseline of 34°C, a stimulation temperature of 43-46°C was reached after 18 sec at 0.67°C/sec. The stimulation temperature plateau was held for 20 sec. Over the subsequent 22 sec, the temperature was dropped linearly back to the baseline. Data analysis was carried out using FEAT (http://www.fmrib.ox.ac.uk) software packages. Z (Gaussianised T/F) statistic images were thresholded using clusters determined by Z>2.3 and a (corrected) cluster significance threshold of P=0.05. Regions of interest (ROI) was manually selected and statistical analyses were performed using an ImageJ. ROI were drawn according to an atlas of the mouse brain (Franklin and Paxinos 1997). BOLD signal intensity values in each ROI were extracted and normalized to the time of baseline (expressed as a percent change from baseline). Drugs The drugs used in the present study were recombinant mouse IL-6 (R&D Systems Inc.), recombinant mouse MCP-3 (R&D Systems Inc.), the selective CCR2 antagonist RS102895 (Sigma Chemical Co.), gp130/Fc (R&D Systems Inc.) and a functional blocking antibody to MCP-3 (30 g/mL, R&D Systems Inc.). IL-6 (1 pmol/mouse), MCP-3 (0.1, 0.3, 1.0 pmol/mouse) and gp130/Fc (10 ng/mouse) was dissolved in sterile PBS containing 0.1% bovine serum albumin. RS102895 (100 ng/mouse) was dissolved in saline containing 1% dimethylsulfoxide (DMSO). Statistical analysis The data are presented as the mean ± s.e.m. The statistical significance of differences between the groups was assessed by one-way analysis of variance (ANOVA) (for pain-like behaviors) or Student’s t-test. METHODS REFERENCES: 1. Hylden, J.L. & Wilcox, G.L. Intrathecal morphine in mice: a new technique. Eur J Pharmacol. 67, 313-316 (1980). 2. Malmberg, A.B. & Basbaum, A.I. Partial sciatic nerve injury in the mouse as a model of neuropathic pain: behavioral and neuroanatomical correlates. Pain. 76, 215-222 (1998). 3. Narita, M., et al. Involvement of spinal protein kinase Cgamma in the attenuation of opioid mu-receptor-mediated G-protein activation after chronic intrathecal administration of [D-Ala2,N-MePhe4,Gly-Ol(5)]enkephalin. J Neurosci. 21, 3715-3720 (2001). 4. Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 72, 248-254 (1976). 5. Narita, M., et al. Enhanced mu-opioid responses in the spinal cord of mice lacking protein kinase Cgamma isoform. J Biol Chem. 276, 15409-15414 (2001). 6. Takeshima, H., et al. The presence of RNA polymerase II, active or stalled, predicts epigenetic fate of promoter CpG islands. Genome Res. 19, 1974-1982 (2009). 7. Tsankova, N.M., Kumar, A. & Nestler, E.J. Histone modifications at gene promoter regions in rat hippocampus after acute and chronic electroconvulsive seizures. J Neurosci. 24, 5603-5610 (2004). 8. Nakajima, T., et al. The presence of a methylation fingerprint of Helicobacter pylori infection in human gastric mucosae. Int J Cancer. 124, 905-910 (2009).