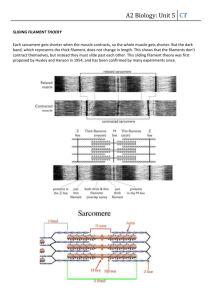
Flagellar Evolution - original paper
advertisement

Evolving the Bacterial Flagellum Through Mutation and Cooption By Mike Gene The bacterial flagellum is a motility structure whose irreducible and specified complexity has been cited as evidence for its design (a good basic description can be found here). Several non-teleological mechanisms have been proposed to explain the origin of Irreducible Complexity (IC), each with its own difficulties [1]. Evolution through cooption is the most commonly cited mechanism, which would involve more than one protein, previously shaped by selection for one function, fortuitously coming together to interact and provide a novel, selectable function. Yet how plausible are such scenarios? Although there is no explanation for the origin of the flagellum in the scientific literature, various non-teleologists from the cyber-community have outlined a vague cooption scenario to explain its evolutionary origin. A detailed analysis of such an scenario might not only help us gage the plausibility of the story, but also help us appreciate what all cooption stories face when attempting to account for the origin of IC machines. The basic cyber-story for the non-teleological origin of the flagellum is as follows: We begin with a type III export system, given that several proteins of the flagellum's basal body are homologous to the secretory machinery of this export system (type III systems secrete various proteins to establish symbiotic relationships with eukaryotic cells). Thus, the flagellum began as a protein secretion system. Next, we hypothesize that some protein, that is normally secreted, is mutated such that it can stick to itself and the secretory system. This forms our proto-filament. Filament formation is not difficult, as a single point mutation in the beta globin gene, responsible for sickle cell anemia, converts soluble hemoglobin into a filament. This filament then could serve the function of anchoring the cell to some other substrate. In fact, if we survey living bacteria, we'll find that there are indeed many different forms of nonmotile filaments that provide benefits to the cell (thus allowing us to propose a selective advantage to this step in the flagellum's evolution). Next, we again invoke cooption, as some other membrane protein somehow associates with the type III/filament system and fortuitously causes it to wiggle in some fashion. This slight movement confers motility to the bacteria, which in turn, is selectively advantageous. From there, mutations are selected that improve the motility function and finally, another set of proteins are coopted to confer the switching of rotation and chemotaxis response. Thus, we have a step-by-step account that involves at least three different functional state: protein export system transformed into nonmotile filament transformed into flagellum. Let us refer to this scenario as the Export-FilamentMotility (EFM) Hypothesis. Yet how well does such an account really explain the origin of the bacterial flagellum? Philosophical Considerations Before beginning a scientific analysis of this scenario, it is worth considering the larger philosophical picture. Many people labor under the impression that IC means "something that even a hefty dose of Darwinian imagination cannot possibly explain." That is, many seems to think that if they (or anyone else) can invent a vague scenario for how the flagellum (or something like it) could have possibly evolved, all the issues brought to the forefront by an IC analysis disappear. But keep in mind that whenever you are dealing with a machine, it is always going to be possible to imagine the various parts existing without the machine, as long as you keep your explanation vague and are free to imagine simpler states with imaginary selective benefits and ad hoc functions. Consider how Ken Miller explains the origin of the bacterial flagellum: But Brown's Mr. Miller contends that the ID argument completely misunderstands how Darwinian evolution works. The various parts of the flagellum weren't destined to serve together as a motor when they first appeared piecemeal in the deep history of bacteria. Instead, the parts served different and separate purposes originally. Only later did they happen to come together to form a motor. Remnants of that process are still visible today in various lines of bacteria, he says. Yersinia pestis, the species that causes bubonic plague, produces a complex structure with 10 proteins closely related to the ones found in bacterial flagella. The deadly bug doesn't use those proteins to move; instead, they help Y. pestis inject its toxins into the cells of its host. Drawing on Mr. Behe's favorite analogy, Mr. Miller says that the various parts of a mousetrap have uses even on their own. Three out of the five components form a handy tie clip. Two of the five can serve as a clipboard. Nature is opportunistic, he says, cobbling together different molecules and reactions for entirely new purposes. [2] What is interesting about this logic is that we already know that the mousetrap was intelligently designed. We also know that it did not first exist as a clipboard, then a tie clip. Thus, while it is logically possible to see the mousetrap as Miller does, that is, as a modified clipboard and tie clip, such perceptions are not tied to history nor the origin of the mousetrap. Thus, coming up with imaginary accounts that tap into our ability to imagine cooptional origins, by itself, is rather meaningless. If we can successfully come up with such explanations where they are known to be false(the mousetrap), how do we know that our ability to do likewise with things like the flagellum are not also inherently flawed? [3] Before we turn our attention to the EFM Hypothesis, it is worth considering something that Julie Thomas posted to talk.origins back in 1997, since this hypothesis has all the characteristics of a just-so story: Here's the recipe for making a just-so story. First, survey the biological world for structures/functions. Find those that seem useful for coming up with a precursor to the system in question and patch them together without much regard for biochemical and/or genetic details. Place the patchwork in an imaginary creature from the distant past that has conveniently gone extinct. Invoke a vague selective pressure that selects for the patchwork and then imagine it is plastic and amenable to further selective modification that just happens to arrive at the system in question. Thus, if all we have is a just-so story that merely reflects our innate ability to imagine non-teleological causes for designed systems, the EFM Hypothesis can hardly be considered a serious rebuttal of the design inference stemming from the flagellum's IC essence. Let's now consider the hypothesis. ASSUMING THE TRANSPORTER FROM THE START The EFM hypothesis begins with the knowledge that a core subsystem of the flagellum is its protein secretion machinery and thus builds around this. That is, to explain the origin of the flagellum, the EFM hypothesis must start with the most complex subsystem that is part of the flagellum (called the type III export system, the machinery invoked by Miller above). However, that flagella have built-in sophisticated transport systems is no surprise from an teleological perspective. With the construction of the flagellum comes a design problem. That is, how does one construct a complex and specific assembly of proteins outside the cell? The bacterial flagellum is quite unlike the eukaryotic flagellum in this regard, as the latter is in direct communication with the cytoplasm and bounded by the plasma membrane. The bacterial flagellum, in contrast, penetrates the cell membrane and wall and is housed outside of the cell membrane. Again, how can it be constructed? It's kind of like putting together a satellite dish on your roof without ever leaving the house. One solution would be to construct the entire flagellum within the cytoplasm and move the whole thing outside. The problem? It would be far too large for such transport. Instead, the design problem has been solved as follows: the flagellum is hollow. This allows bacteria to construct flagella from the bottom up. To facilitate this construction, a significant chunk of the flagellar machinery functions as a protein secretion/export machine that channels components through the hollow space to distal ends where they attach (resulting in piecemeal growth of the flagellum). The interesting thing is that the mistakes in construction at any point actually shut down the expression of gene products that would be used in the later stages of flagellar construction, thus imparting a built in quality control mechanism for flagellar synthesis. Nevertheless, the export machinery solves a design problem entailed in the cell constructing a flagellum. Thus, we can see that an integral component of a flagellum is its modular export machinery needed to construct and maintain the flagellum. It's a clever solution to a clear design problem. In the past, I have asked ID critics just what would the flagellum look like if it was not designed. After all, if you pay attention as I do, they commonly argue that this and that does not look like it was designed. Nevertheless, the critics have not answered this question. Yet if the transport/secretion system is a logical ingredient that solves a design problem entailed in making the flagellum, and the flagellum was indeed designed, those looking for non-teleological explanations would misinterpret the significance of such a subsystem and mistakenly impose an historical interpretation on an engineering solution. Now, if someone wants to start this story with "any ol' transporter," I'm afraid that's not good enough. Remember, that we need to explain the origin of the bacterial flagellum (not some "flagellum"). That means we need to account for the flagellum's type III export machinery, which includes flhA, flhB, fliR, fliQ, fliP, fliI, and more. All of the other bacterial transport/secretion systems cited to support the EFM hypothesis merely illustrate that the majority of transport/secretion systems are dead-ends from a flagellar perspective, as none of them have spawned a eubacterial flagellum, despite them all being equally good starting material at this point in the EFM hypothesis. It's important to keep in mind that there are indeed dead-ends in evolution. For just one example, consider the inverted retina of the vertebrate eye. As Ken Miller explained elsewhere: Evolution, which works by repeatedly modifying preexisting structures, can explain the inside-out nature of the vertebrate eye quite simply. The verterbate retina evolved as a modification of the outer layer of the brain. Over time, evolution progressively modified this part of the brain for light-sensitivity. Although the layer of light-sensitive cells gradually assumed a retina-like shape, it retained its original orientation, including a series of nerve connections on its surface. Evolution, unlike an intelligent designer, cannot start over from scratch to achieve the optimal design. [4] Evolution cannot start over and must work with what it is handed. We've heard much about the suboptimal retina. The reason why random mutations coupled to nature selection cannot "fix" it is because it is essentially hardwired in early embryological development. In other words, "choices" made early on in a progressive march of construction not only lack foresight, but constrain what can happen next. The blind watchmaker can box himself in with each new choice. Thus, it would seem that we'd need to establish two things for the plausibility of the EFM hypothesis. 1. Would "any ol' transporter" really do? That is, could we take the framework of any ol' transporter and put the type of flesh on it that is exhibited by the bacterial flagellum? Or would most of these transporters be dead-ends in the sense that their transport mechanism entails a constraint that would prevent the evolution of something like the flagellum-aswe-know-it? Again, this latter point seems to be the case since none of the other transport systems evolved something comparable to flagellum among eubacteria. 2. Is there any reason to think the type III export system, complete with the ancestors of flhA, flhB, fliR, fliQ, fliP, fliI and others, existed as a "cooptable part." Thus far, the answer is no, as there are good reasons to think the type III system evolved from preexisting flagella. a. The bacterial flagellum is found in both mesophilic and thermophilic bacteria, grampositive and gram-negative, high GC and low GC content bacteria, and spirochetes. Type III systems seem to be restricted to a few gram-negative bacteria. That is, if we look at the sequenced genomes from the various groups cited above, we can find the genes for the bacterial flagellum but not the type III system genes. b. Independent evidence suggests the type III system is recent. It is not only restricted to gram-negative bacteria, but to animal and plant pathogens. In fact, the function of the system depends on intimate contact with these multicellular organisms. This all indicates this system arose after plants and animals appeared. In fact, the type III genes of plant pathogens are more similar to their own flagellar genes than the type III genes of animal pathogens. This has led some to propose that the type III system arose in plant pathogens and then spread to animal pathogens by horizontal transfer. c. When we look at the type III system its genes are commonly clustered and found on large virulence plasmids. When they are in the chromosome, their GC content is typically lower than the GC content of the surrounding genome. In other words, there is good reason to invoke horizontal transfer to explain type III distribution. In contrast, flagellar genes are usually split into three or more operons, they are not found on plasmids, and their GC content is the same as the surrounding genome. There is no evidence that the flagellum has been spread about by horizontal transfer. d. It's much easier to envision the evolution of the type III system from flagella than vice versa. For starters, evidence has surfaced that the basal body of the flagellum already works to secrete proteins other than the flagellar proteins, including virulence factors. Thus, the basal body is already poised to evolve into a type III system from the start. Evolution apparently would only have to duplicate and tweak the type III virulence protein secretion activity already existing in flagella. . In my opinion, this view is far more parsimonious than to propose that something like the type III system evolved long ago, was lost by all bacteria but gram-negative animal/plant pathogens and then was used to evolve the flagellum so that horizontal transfer could spread flagella far and wide (despite the lack of evidence for such transfer). Thus, it should not be surprising that the scientific opinion has been converging on the notion that the export machinery evolved from the flagellar machinery [5-7]. Is there any evidence that supports transporting this system, or something like it, back in time? The type III system is one of at least four different bacterial protein transport systems. And it appears to be the most complex of the bunch. The key here is that the type III/flagellar cytoplasmic export system does not show clear homology with any of these other transport systems. But we also know that evolution builds on and modifies what already exists rather than create de novo. Thus, if these other transport systems were already in place (and they probably were), why didn't evolution simply build on one of the simpler versions rather than create a whole new method of protein secretion de novo? The type III-from-flagellum scenario better fits with what we know about evolution - that it uses what already exists rather than inventing de novo. Thus, not only is there no evidence to support putting this transporter (or something closely homologous) back in pre-flagella days, there is reason to think it wouldn't be there. Another fundamental problem with the first step of the EFM hypothesis is that it assumes IC to explain IC. Consider that the flagellum is composed of about 20 different proteins. Of those twenty, ten are homologous to the type III export machinery. Thus, this hypothesis begins its attempt to explain the origin of a 20-part IC system by assuming the existence of half of it (10 parts). Let's briefly consider the 10 parts of the type III export machinery, where I'll use the flagellar gene names. There is FliI, an ATPase that is anchored to cytoplasmic face of the inner membrane. It may provide energy for the synthesis of the export machinery or transport of secreted proteins. It is thought to capture proteins in the cytoplasm for transfer to the secretory apparatus. There are several proteins that span the inner membrane and probably make up the protein conducting channel, including FlhA, FliP, FliQ, FliR, and FlhB. There is FliF, which in flagella, form the MS ring. And there is FliN and FliG, which in flagella, make up the C ring (located beneath the MS ring). Finally, there is FliH, with an unknown function. Whether all 10 proteins are required for type III transport is unknown, but it appears most are essential: Several proteins essential to secretion including LcrD, YscD, R, S, T, and U, are known or predicted to reside in the inner membrane (10,12,48-50). At the outer membrane, only one protein, YscC (48), and two lipoproteins, YscJ and VirG (51,52), appear essential for proper secretion. The roles and subcellular locations are not known for several more essential proteins, YscE, F, G, I, K, and L (9,48,51). How all of these proteins interact with one another to form the secretion apparatus is not yet understood. It is clear, however, that correct assembly of the apparatus is required not only for secretion, but also for normal synthesis of effector molecules (47,48). If one component of the export machinery is missing, production of the effector molecules is altered. [7] LcrD, YscD, R, S, T, and U are FlhA, FliG, FliP, FliQ, FliR, FlhB, respectively. FliI is also thought to be essential. In other words, the EFM hypothesis fails to account for the origin of the protein secreting channel itself, which is made up of several independent proteins. There is yet another interesting aspect to all this. Since evolving from some flagellum, the type III transport system appears to have lost its ability to engage in rotary transport. The flagellar motor is composed of five proteins: MotA, MotB, FliG, FliN, and FliM. We'll discuss this more below, but right now it is worth pointing out that the type III systems have no homologs for MotA, MotB, or FliM. The Mot proteins are essential components of the motor, as they are membrane proteins that fulfill two functions: they transport ions to provide the energy for rotation and serve as the stator against which the rotor (FliG, FliN, FliM) moves. What's more, the type III rotor components have significantly changed. The type III homolog of FliN shares sequence similarity only with in its Cterminal 80 amino acids. And the sequence similarity between the FliG homologs are almost non-existent. Furthermore, there have been significant changes in FliF. FliF forms the MS ring (the "mounting plate"), which is associated with and above the C-ring composed of FliG, M, and N. FliF in flagella is composed of 500+ amino acids, but in the type III homolog, both the C- and N-terminal domains thought to be involved in forming the MS ring are missing. All that is left in common between them is a central region of about 90 amino acids. Here we find another reason to recognize the significance of the flagellum-to-type III system evolution. Type III systems have apparently lost their ability to rotate. Thus, we can't think of type III systems as something pre-adapted to rotate, as all the rotary information has been lost. To argue that the type III system could reacquire the ability to rotate, as the flagellum does, is to essentially violate Dollo's Law, which states: "evolutionary change manifested at any level higher than the genetic is irreversible, and that anatomical structures or functions once lost cannot be regained." [8] Yet by proposing that the flagellum once existed as a type III system and later acquired the ability to rotate is not hardly any different that proposing type III systems could reacquire the ability to rotate and violate Dollo's Law. To summarize, the problems with the first step in the EFM are as follows: There are good design reasons for building the flagellum around a transport system and attempts to assign historical significance to this are completely unsupported. We have the ability to imagine simpler initial states for designed systems where our imagination does not reflect reality. Thus, the mere ability to imagine simpler states is rather meaningless. The data suggest that not any transporter is good enough for evolving something like the bacterial flagellum. This is important because the EFM never addresses the origin of the transporter itself. It seems quite plausible that selection pressures for evolving a good transporter would steer the system away from being preadapted to form something like the flagellum (which is why bacterial flagella are monophyletic). There is no evidence that anything like the type III system predated the flagellum. The type III system itself most likely evolved from a flagellum. Because it is known that evolution borrows, rather than invents de novo, proposing the de novo evolution of a type III transport system among bacteria that already had several transport systems in place makes little sense. The type III system itself is IC, perhaps with ten IC components. No attempt is made to explain its flagella-independent origin. In light of the evolution of the type III system, proposing a type III-to-flagellum evolution appears to violate Dollo's Law. NEXT: In the next essay, I will consider the cooptional addition of the filament to determine just how grounded this just-so story is in biology. Citations 1. IC ReVisited, TeleoLogic, no. 7 2. Monastersky, R. Seeking the Deity in the Details. Chronicle of Higher Education, 12/21/2001. 3. At this point, one might want to argue that the mousetrap does not self-replicate. But then, neither does the flagellum. Both are assembled from parts. The difference is the flagellum is assembled by a cell, while the mousetrap is assembled by a factory. Are we therefore to believe that once upon a time, a clipboard factory made a mistake in its production line and produced tie clips? Then, the newly spawned tie-clip factory later made a mistake in its production line and began making mousetraps? 4. Miller, K. 1994. Life's Grand Design. Technology Review 97:24-32. 5. Nguyen L, Paulsen IT, Tchieu J, Hueck CJ, Saier MH Jr. 2000. Phylogenetic analyses of the constituents of Type III protein secretion systems. J Mol Microbiol Biotechnol Apr;2(2):125-44. 6. Stephens C, Shapiro L. 1996. Delivering the payload. Bacterial pathogenesis. Curr Biol Aug 1;6(8):927-30. 7. Molecular Mechanisms of Bacterial Virulence: Type III Secretion and Pathogenicity Islands. 8. Dollo's Law ID THINK Evolving the Bacterial Flagellum Through Mutation and Cooption Part II Evolving the Bacterial Flagellum Through Mutation and Cooption: Part II Let us now turn our attention to the hypothetical addition of a filament to a protein export system: Next, we hypothesize that some protein, that is normally secreted, is mutated such that it can stick to itself and the secretory system. This forms our proto-filament. Filament formation is not difficult, as a single point mutation in the beta globin gene, responsible for sickle cell anemia, converts soluble hemoglobin into a filament. This filament then could serve the function of anchoring the cell to some other substrate. In fact, if we survey living bacteria, we'll find that there are indeed many different forms of nonmotile filaments that provide benefits to the cell (thus allowing us to propose a selective advantage to this step in the flagellum's evolution). ADDING THE FILAMENT Let us first consider filament formation with sickle cell hemoglobin, as this example highlights problems with such a cooption scenario rather than supporting it. Hemoglobin is normally a globular, soluble protein. In sickle cell anemia, a mutation has occurred in the sixth position of beta globin that substitutes a hydrophobic valine residue for a charged glutamate. This results in a conformational change when the protein is not bound to oxygen that exposes the newly created oily patch (thanks to valine) on the surface. Thus, individual hemoglobin proteins can now stick together in a chain-like fashion because every protein has two oily patches (due to there being two copies of beta globin per hemoglobin molecule). Yet there are some characteristic features of HbS (sickle cell hemoglobin) filaments that make them much different from any of the surface filaments the EFM hypothesis may allude to, including the flagellum. HbS filaments are homogenous, being composed only of chains of hemoglobin. Bacterial surface structure filaments are heterogeneous and ordered, as we shall explore some examples. HbS filaments form inside the membrane of the red blood cell (RBC). In fact, something like 30-40% of the RBC's volume is filled with hemoglobin. This crowding ensures that the oily patches of the HbS proteins are likely to find each other and form stable interactions. None of this applies to the surface structure filaments of bacterial, which form on and outside the cell's membrane. HbS filaments are solid, while the surface structure filaments are hollow. This is especially relevant in the case of the bacterial flagellum, as its hollow core is essential to its construction. When we consider the differences outlined above, it should be clear that any filament formation associated with the bacterial structures is going to be quite different from the process of HbS filament formation, indicating that any claim of filament formation being easy for a protein is misleading in this context. It is understandable that the EFM hypothesis keeps the cooption event so vague, as attempting to be a little more specific would only highlight the inherent problems associated with such musings. For example, it proposes a single mutation that does two things, causing the exported proteins to stick to themselves and causing the exported proteins to stick to the outer components of the export machinery, is necessary to bring about some change that is even vaguely selectable. That is, we assume such simultaneous changes because if they are separated, export proteins that simply stuck to themselves upon secretion would simply float away as multimers and export proteins that stuck only to the export machinery would merely decorate it, perhaps hindering transport of other material, and not produce anything that resembles a filament. But in order to have these simultaneous changes, one might expect the mutation to be rather non-specific. And this poses a huge problem for the EFM hypothesis. What is to prevent the exported proteins from aggregating before they are transported? What's to prevent them from sticking to something else not in the basic story? What's to prevent them from being degraded? And what's to prevent them from clogging up the transport channel during any stage of the transport process? And what if this mutant export-protein polymerized like HbS filaments and formed a solid core? Then, we'd seal off the export machinery, which presumably is important for other reasons. Thus, it would seem the EFM hypothesis is relying on deleterious mutations to evolve his filament, unless of course, it is relying on a special mutation that just happens to perfectly fit the story. And then I can't help but wonder just what type of advantage there is to having some protein filaments globbed onto the export machinery. One might argue that there are many uses for nonmotile filaments as seen in extant bacteria, such as adhension and conjugation. Yet we can rule out things like conjugation as such functions are unlikely to have been carried out in the very simple filament imagined by EFM hypothesis (conjugation, for example, requires another story about IC machines). I suppose adhering to a surface could be useful, although in most cases, the surface is usually another cell. But that brings up another point about this special mutation. Not only is it supposed to allow for the exported protein to stick to themselves, without aggregating into some clump prior, during, or just after transfer, and it is supposed to allow for the exported protein to stick to the export machinery without clogging it, but now the end of this filament must also stick to something else. A mutation that does three good simultaneous things without causing any harm. Like I said, it's special. Nevertheless, it would seem even if we had a special mutation, the export machinery is now attached to some filament, which would seem to cause some form of interference with the other material the system was previously selected to transport. Thus, regardless of the mysterious advantage associated with attaching a proto-filament to the export apparatus, we ought not ignore the disadvantages associated with gunking it up. Yet the EFM hypothesis does exactly this. Non-telic transition stories typically have to keep things simple and sloppy like this. But in doing do, one wonders if they also have to abandon biology. Let's consider what is known to be involved in the simplest and most common form of filament formation in bacteria. Let's consider the P pilus as a model system to see how close it is to the EFM hypothesis. The P Pilus The P pilus is a very thin filament, whose outer diameter is only about 7 nm with a hollow core about 2 nm in diameter. The rod is thicker near the membrane and thins as it nears the tip. It functions as an attachment organelle, that is, it can reach out and anchor bacterial cells to other cells. The end of the filament has a protein that specifically binds to certain sugar molecules found on kidney cells. Although the P pilus is among the simplest of attachment filaments, it is encoded by 11 genes. The filament itself is a heterogeneous structure. The primary subunit is PapA (it forms the thicker rod near the membrane). But as we get near the tip, we find another protein, PapE, forms the thinner filament.. At the very end, is PapG, the specific adhesin that binds to sugars on other cells. PaPG binds to PaPE through an adaptor protein, PapF. And PapE binds to PapA through another adaptor protein, PapK. Thus, the pilus itself is composed of five different proteins that are assembled in a fixed order (PapA - PapK PapE-PapK-PapG, proximal to distal). How is this pilus synthesized in such an orderly fashion? Like most other pili and adhesive organelles, it starts with the highly conserved usher/chaperone pathway. And here is where things not only get interesting, but also begin to look very different from the simplistic account of filament formation assumed by the EFM hypothesis. It all begins with the Sec export machinery found in the cytoplasmic membrane: Protein translocation across the bacterial cytoplasmic membrane has been studied extensively in Escherichia coli. The identification of the components involved and subsequent reconstitution of the purified translocation reaction have defined the minimal constituents that allowed extensive biochemical characterization of the so-called translocase. This functional enzyme complex consists of the SecYEG integral membrane protein complex and a peripherally bound ATPase, SecA. Under translocation conditions, four SecYEG heterotrimers assemble into one large protein complex, forming a putative protein-conducting channel. This tetrameric arrangement of SecYEG complexes and the highly dynamic SecA dimer together form a proton-motive force- and ATP-driven molecular machine that drives the stepwise translocation of targeted polypeptides across the cytoplasmic membrane. Recent findings concerning the translocase structure and mechanism of protein translocation are discussed and shine new light on controversies in the field. [1] However, since this four-part machine is ubiquitous and has many uses other than helping form filaments, let's grant its existence and deal with it by itself as a separate topic some other day. Also, at this point, I should mention that we're discussing gram-negative bacteria, which have two membranes. The inner membrane is a typical cytoplasmic membrane and the outer membrane is more porous (due to many barrel-shaped protein pores that filter out large material but allow smaller things like sugars and amino acids inside). The space between the two membranes is called the periplasm. Transport via the Sec pathway dumps material into the periplasm. The trick for the bacteria is to grow this into a filament that penetrates the outer membrane in a coordinated manner. So how do cells make P pili? First, you export all the pilus subunits into the periplasm using the sec-machinery. The proteins are threaded through the sec-machinery in an unfolded state and most refold in the periplasm. And therein lies the problem, as the pilus subunits easily form insoluble aggregates (or clumps) in the periplasm through hydrophobic interactions. To prevent this, we need to invoke another component, a special chaperone encoded by PapD. PapD does two things - it binds to the pilus subunits after they are pumped into the perisplasm and prevents them from clumping with each other and also helps the pilus subunits to fold into their proper conformation. In fact, the pilus subunits are not stable as monomers and exist either as bound to the chaperone or as bound to each other as part of the filament. The manner in which the chaperones carry out their function is far more elegant than anyone assumed, employing something that is now called "donor strand complementation" (DSC). The 3-D structures of PapD complexed with PapG (the adhesin on the tip) and PapK (one of the adaptors) have been solved. PapD forms a boomerang-shaped protein with two immunoglobulin-like (Ig-like) domains (a structure composed of layers of antiparallel beta sheets). The N-terminal end of PapK is also an Ig-like domain, but it lacks a Cterminal beta sheet that normally contributes to the hydrophobic core of the domain. This produces a cleft that exposes the hydrophobic core, which is what makes it so sticky and prone to aggregation by itself. The chaperone PapD masks this exposed region in a most fascinating manner - it donates one of its beta strands to complete the Ig-domain in PapK (Fig 1). But it does so in an atypical fashion, as the beta strand it donates runs parallel, not antiparallel, with its neighboring strand. Thus, PapD provides at least two essential functions captured in one very elegant act - by donating one of its beta strands, PapD simultaneously prevents aggregation of PapK while providing the missing steric information for proper folding of PapK. Fig 1. Modified from [2] And what this means is the folding of pilus subunits is IC. By themselves, the subunits don't fold properly and are unstable. The steric information for proper folding is not found in a single amino acid chain or gene, but in two distinct chains/genes. And By itself, PapD has no function. Clearly, the simplest known filament is far more sophisticated than the filament imagined by the EFM hypothesis (i.e., biology is not as simple as it assumes). What happens next? The pilus subunit-chaperone complex interacts with a protein channel on the outer membrane, PapC (also known as the usher). The channel is large enough to accommodate the tip of the filament, but not the rod. The actual mechanism of incorporation is being worked out, as the chaperone somehow hands off the pilus subunit to the usher for incorporation into the growing filament. Interaction between the usher and chaperone-pilus subunit does not result in the chaperone-subunit complex breaking apart, thus the mechanism of handoff is also probably quite complicated and sophisticated. But there is one more feature to the story worth mentioning. The pilus subunits themselves are thought to form a filament through a donor strand complementation-like mechanism. Each pilus subunit has an N-terminal extension that does not contribute to its own folding. By itself, it is a disordered strand. However, it has been proposed that this N-terminal extension from one subunit (let's call it A) displaces the displaces the donated chaperone strand associated with another pilus subunit (B). This N-terminal strand would then form a beta strand that runs in an antiparallel direction and complete the Ig-domain of its neighbor in a typical fashion.(Fig 2) Again, the steric information for the Ig-domain of subunit B is supplied from subunit A. This mechanism is called donor strand exchange. And the result is that the filament is made by linking subunits, where each subunit contributes a strand to perfectly complete the fold of its nearest neighbor. Fig 2. Modified from [2] Thus, it should be clear that some ad hoc notion of an export protein sticking to itself and sticking to the export apparatus to form a filament does not reflect the biology of the simplest known pilus. Life is much more sophisticated than this. Thus, all the examples of simple, nonmotile filaments in bacteria provide no obvious support for the EFM speculation. As if having your supporting evidence shown to be irrelevant was not bad enough, there are more problems. For example, let's imagine that with enough luck, somehow a P-piluslike materializes. After all, such pili are the most common. And therein lies the problem, because while the P-pilus makes a great attachment organelle, it's probably a dead-end if one wants to evolve a flagellum. For one reason, the P-pilus has not been observed to secrete proteins. This could be because the channel is so small . Or it might have something to do with the energetics of the system, as P pili formation is independent of cellular energy. It's not surprising that the P-pilus looks very different from the bacterial flagellum (or even things like type IV pili). Finally, there is yet another fact that suggests flagella did not arise in the manner that the EFM proposes. Whether we're talking about simple type I pili or more complex type IV pili, what they all share in common is being built from the bottom-up. The flagellar filament, in stark contrast, is built from the top down. And the manner in which this done is yet another amazing story in microbiology. How amazing? Robert Macnab is an expert on the flagellum and has been working on them his whole life. As such, you might expect him to be used to the complexity and sophistication of the flagellum. Yet he reacted by noting that this mechanism is " a much more sophisticated process than any of us could have envisaged."[3] In fact, consider how this was reported: The latest technical discoveries in flagella fascinate biologists such as Robert Macnab, a professor of molecular biophysics and biochemistry at Yale University who also studies flagella. He marvels at how organisms as simple as bacteria have evolved such complex methods to develop propelling features, especially since motility in bacteria is not directly necessary for survival, like DNA replication or protein synthesis. "We think it would not be possible for the system to work with any significantly lower complexity." [4] So let's have a look to see how well the EFM hypothesis' filament formation story anticipates the actual mechanism bacteria use to form filaments. Flagellar Filament Formation Shigella are nonmotile pathogens. Even though Shigella do not express flagella, they do possess the flagellar operons, suggesting this nonmotile state was recently acquired. Four strains were recently analyzed, showing that loss of flagella has occurred independently.[5] In two strains, the only thing missing was fliD, the gene that codes for the protein that caps the filament. What happens if you don't have fliD is that no filament forms? As Ikeda et al. explain, "A fliD-deficient mutant becomes non-motile because it lacks flagellar filaments and leaks flagellin monomer out into the medium." [6] FliD is not merely a regulator or aid, but an essential component for filament formation. To understand why, let's consider the research results that fascinated Macnab and others.[7] The fliD gene products form a five-member pentagon-shaped ring that caps the hollow filament formed by flagellin subunits. Each member of this pentamer has a leglike extension that points downward and interacts snuggly with the filament. However, there is a symmetry mismatch between the cap and the filament. The cap is formed from five protein subunits, but the helical end of the filament itself is formed from 5.5 flagellin subunits. Macnab explains the significance of this as follows: " When one protein of the cap pentamer is at the dislocation point (think of a split washer), it will be in a very different environment from the other four members of the pentamer." [3] In other words, a significant crevice is associated with the cap and end of the filament. And it is proposed that the next flagellin subunit that gets added to the filament is added to this crevice. The addition of the new flagellin subunit is then coupled with the cap itself rotating along the filament axis to open up a new adjecent crevice. As Macnab suggested, think of the cap as a split washer (where the center is filled) sitting on the end of a hollow tube. Individual flagellin proteins travel down to the tube to be added at the tip. The flagellin then gets placed into the space of the split washer, the washer turns, and opens up a new space. Thus, you can envision the cap spinning around, inserting new flagellin monomers one-at-a-time. (Fig 3) Fig 3 (adapted and modified from [7]) [The yellow blocks represent flagellin. Newly added flagellin molecules are shown in violet. As the cap turns, one of its legs exposes an empty slot (shown in the picture second from the left). This slot is the site for the next addition of flagellin. ] Duane Salmon once estimated that the growth rate of the filament to be about 50 flagellin units/sec.[8] Since there are ca. 5 subunits per turn of the helical filament, this suggests that the fliD cap rotates about ten times every second as it incorporates about 50 flagellin subunits. What's most relevant about this is that the C-terminal and N-terminal ends of flagellin subunits are unfolded as they travel down the hollow filament tube, as the folded protein has a significant kink in its middle that would prevent transport through the tube. As Macnab notes, "large conformational changes would be required in the monomers before they could be added to the filament tip." Thus, the fliD cap also does not simply provide a passive, mobile slot to insert flagellin subunits. It also helps flagellin fold. In other words, the cap is a chaperone. Thus, the flagellar filament is built in a way that is similar to P pili and quite different from HbS filaments; the flagellin units do not "self-assemble," they are assembled by a processive chaperone at a rather impressive rate. Things get even more interesting when one considers that just below the cap, the filament cavity is expanded such that its cavity is about twice the size of the central channel that runs through the rest of the filament. It is suggested that this might be the site in which flagellin folds in a manner that is analogous to the folding that occurs in the GroEL chaperonin in the cytoplasm. The parallels are interesting. GroEL is capped by GroES to form a closed chamber, while FliD also functions as a cap to form a closed chamber. It is suggested the filament chamber can house one flagellin monomer at a time, which is exactly how GroEL works. Yet there are a couple of significant differences that probably stem from the fact that GroEL is a generic chaperone chamber that functions only to fold a diverse set of proteins, while the chamber at the distal end of the filament folds and incorporates only one protein, flagellin. The first difference is that GroEL requires energy in the form of ATP hydrolysis that alters the volume of the chaperonin. It is intriguing to speculate that the folding chamber at the end of the filament also undergoes cycles of volume changes associated with the rotation of the cap and insertion of a new flagellin filament. In such a case, the energy could be derived from the winding coupled to favorable protein-protein interactions associated with assembling flagellin subunits into the filament. Secondly, the filament chamber would cycle much faster that GroEL. The typical GroEL cycle lasts 15 sec. The filament, on the other hand, is incorporating 50 subunits/sec. That's folding individual monomers every 0.02 seconds, which is 750 times faster than GroEL. There are several clues that point to design here. 1. Flagellin/fliD and GroEL/GroES are not homologous. Yet if the flagellin/fliD chamber functions as I suggest, we have another system whose sophisticated mechanism is related in a logical fashion (another example would be in the similar proofreading mechanisms of DNA replication and attaching amino acids to tRNA). 2. FliD and flagellin form an IC relationship. FliD has no other basic cellular function apart from forming the filament. Flagellin too has no other basic cellular function apart from forming the filament. And both are needed to form the filament. 3. As suggested, there seems to be only enough room for one flagellin monomer to fit into the chamber and fold. If this is essential, we have another IC-like interaction. Flagellin must be first unfolded to transport through the channel. But it must also be folded again to be incorporated into the filament. If this second folding event depends on the distal chamber, then two independent events must be carefully coordinated to construct the filament. And there is one more interesting twist on all of this. There is suggestive evidence that the hook-associated proteins, those that attach the filament to the basal body and the fliD cap itself, may be chaperoned through donor-strand complementation. Specifically, there are two chaperone proteins that specifically interact with the C-terminal ends of the hookassociated proteins and cap and prevent their premature aggregation. Thus, just as there is a mini-IC relationship with flagellin and the cap, the cap and hook proteins may also share an IC relationship with their specific chaperones. Again, we would see the basic conceptual strategy in protein folding and assembly as seen independently in the P pilus. And the "self-assembly" is highly regulated - a chaperone helps assemble the hook, another chaperone helps assemble the cap, and the cap assembles the filament. In other words, and here is the interesting point, we will soon begin to make a strong argument that assembly of the flagellum itself is IC. To sum this section up, let's consider more problems inherent in the EFM hypothesis The EFM hypothesis is divorced from biological reality, as the formation of the simplest filaments (the p pili) is far more involved (at its core) than a protein simply sticking to itself. The EFM points to other filaments that employ bottom-up construction to explain the top-down construction of the bacterial filament. It is not clear that a transport system, by itself, is "preadapted to form a filament." Even if it is true that secretion systems are preadapted to form a filament, such "preadaptation" may very well steer a forming structure away from the fitness peak associated with a flagellum-like structure. For example, the most common filaments do not transport proteins, probably because they are too small and lack sufficient energy sources: "Thus, the chaperone/usher system might not be able to adapt for secretion of soluble proteins." And there is no reason, according to the EFM hypothesis, that the filament must be hollow. One might claim there are lots of uses for nonmotile filaments currently in use by living bacteria. Yet how many have gone on to become rotary propulsion units? The bacterial filament itself, along with its assembly process, is IC. It is fundamentally more sophisticated and complex than anything foreshadowed by the EFM hypothesis, indicating again that this hypothesis is divorced from biological reality. NEXT: In the next essay, I will consider the cooptional addition of the motor to determine just how grounded this just-so story is in biology Citations 1. Manting EH, Driessen AJ. 2000. Escherichia coli translocase: the unravelling of a molecular machine. Mol Microbiol Jul;37(2):226-38. 2. Donor Strand Exchange 3. Science 290, p. 2087 4. Bacteria create natural nanomachines 5. Al Mamun AA, Tominaga A, Enomoto M. 1997. Cloning and characterization of the region III flagellar operons of the four Shigella subgroups: genetic defects that cause loss of flagella of Shigella boydii and Shigella sonnei. J Bacteriol 179(14):4493-500 6. Ikeda T, Oosawa K, Hotani H. 1996. Self-assembly of the filament capping protein, FliD, of bacterial flagella into an annular structure. J Mol Biol 259(4):679-86. 7. Yonekura K, Maki S, Morgan DG, DeRosier DJ, Vonderviszt F, Imada K, Namba K. 2000. The bacterial flagellar cap as the rotary promoter of flagellin selfassembly. Science 290(5499):2148-52. 8. Duane ID THINK Evolving the Bacterial Flagellum Through Mutation and Cooption: Part III ID THINK Evolving the Bacterial Flagellum Through Mutation and Cooption Part II Evolving the Bacterial Flagellum Through Mutation and Cooption: Part III Let us now turn our attention to the evolution of motility. The EFM hypothesis essentially proposes: Next, we again invoke cooption, as some other membrane protein somehow associates with the type III/filament system and fortuitously causes it to wiggle in some fashion. This slight movement confers motility to the bacteria, which in turn, is selectively advantageous. At this point, the EFM hypothesis becomes so vague and so speculative that it borders on the vacuous. Such assertions are without any independent support and have all the appearance of an ad hoc explanation invoked to rationalize a previously held belief in the cooptional origin of the bacterial flagellum. Nevertheless, let us consider current thinking about the flagellar motor and then come back to consider such claims in that light. The Flagellar Motor Parts As of today, we have a fairly good handle on the components of the bacterial motor: Mot A, Mot B, FliG, FliM, and FliN. The motor itself is broken down into two basic components - a stator and a rotor. The rotor is the component that turns and the stator is an adjacent component against which the rotor turns. The rotor is composed of FliM, FliN, and FliG. The proteins form a C-ring structure at the base of the flagellum just underneath the cytoplasmic membrane. (Figure 1). Figure 1. Adapted from [1] This C-ring is composed of about 25-45 copies of FliG, about 35 copies of FliM, and around 110 copies of FliN. [2] It actually has three functions. [3] First, it plays an essential role in flagellar assembly. Secondly, it is part of the motor. And third, it is also part of the switch complex that mediates clockwise and counterclockwise spinning. A recent study from the March 23,2001 issue of Science helped to clarify the assemblyessential role of the C-ring. Apparently, it functions as a "quantized measuring cup" for the hook monomers. The current model is that the interior chamber of the C-ring is loaded with about 120 hook monomers (where FliM, FliN, and FliG each have four binding sites for the monomer) and these are then secreted en bloc to form a hook of a distinct length (helping to explain how the flagellum controls the assembly of its hook, as the heterogeneous natural of the hook/filament is often overlooked by some). After the hook monomers vacate the C-ring, another protein enters the chamber and converts it from a hook-secretor to a flagellin-secretor. When it comes to motility, it appears that FliG plays the primary role, as FliM and FliN are more involved in switching. FliG is protein that is approximately 331 amino acids long. It is thought be directly involved in the generation of torque as a consequence of its specific interactions with the stator. The stator is composed of motA and motB. These are both membrane proteins, where removal of either one abolishes motility. MotA has four membrane-spanning regions and most of its bulk is found on the cytoplasmic side of the membrane. MotB has only one membrane spanning domain and most of its bulk is in the periplasm, where it is anchored to the underside of the bacterial cell wall. Together, they form the torque generating units, as the not only form a stationary structure against which the rotor can move, but conduct sodium ions or protons (depending on the flagellum) from the periplasm to the cytoplasm and this ion/proton flow generates the spinning of the C-ring. There are at least 8 copies of the motA/motB torque generators that surround the C-ring (keep in mind that Figure 1 is a cross section through the flagellum). Torque Generation Models The actual mechanism is torque generation is poorly understood. Let me briefly describe three models. The first model, the "proton turbine model," proposes electrostatic interactions between motA/B and FliG, as can be shown by the right illustration in Figure 2 (keeping in mind only one torque generator is shown). Figure 2. Adapted from [4] This model proposes that the flow of protons or sodium ions interact with carefully positioned charged residues on the FliG component of the C-ring, creating a dynamic electrostatic field that moves the rotor. The second model is sometimes called the "turnstile" (shown in the illustration on the right). Protons or ions enter the motA/B complex and are passed on to specific components of the rotor. Yet the rotor must spin to again pass on the protons/ions for entry into the cytoplasm. The third model (not shown) is called the "water turbine model." [5] In this model, protons or sodium ions bind to residues on motA/B. Normally, the protons/ions are complexed by a surrounding sphere of water molecules. Their binding to amino acid residues causes the water molecules to be vectorially ejected: "the binding of protons (or Na+ ions) to specific groups on MotA leads to the vectorial ejection of water molecules tangentially to the C-ring, thus causing its rotation."[5] In this model, the water molecules become an active participant in rotary motion. As of today, the first model, built around electrostatic interactions, appears most popular. What all models share in common is the theme of specificity, whereby specific interactions between the stator and rotor are required to elicit rotation. For example, Electrostatic interactions are weaker in water than in a less polar milieu. They could still exert significant forces, however, particularly if the interacting groups are positioned in the motor so as to ensure that they will approach each other closely at some step(s) in motor rotation. Also, water might be partially excluded from the rotor-stator interface, which would make the interactions stronger. (emphasis added) [6] Or consider FliG. The torque generation function associated with this protein is restricted its C-terminal domain. Random mutations were introduced into the gene coding for this protein and yielded a set of mutants with flagella that did not rotate. In fact, even if the mutant protein was overexpressed, motility was not restored. All but one of these mutants involved the loss of a hydrophobic amino acid and were found to make the proteins subject to degradation. [7] This likely means that these mutations altered the conformation of FliG such that its charged residues directly involved in torque generation were no longer properly positioned. The mutations involved residues at positions 234, 237, 249, 252, 257, and 306. Remember that a mutation in any one of these sites resulted in complete loss of motility. In addition, small deletions also had the same effect: deletions of residues 280-285 and 292-295. With this in mind, let us return to the EFM hypothesis. The Fortuitous Interaction The EFM hypothesis envisions some ill-defined fortuitous interaction between some unknown ion channel and the basal body of the non-motile filament. Then somehow, motility spontaneously emerges and selection takes over from here. Once again, pure chance must bring about the new function. But how likely is this? First, I'm having a hard time envisioning this. The rotor must have access to the proton/ion through the ion channel. What type of fortuitous change is going to pry open this ion channel and then glom it onto the proto-rotor? It would seem that of all the ways to mutate an ion channel, such a change would represent only a very small minority of all possible changes. In fact, this very same theme repeats itself with all necessary parts of this fortuitous interaction. Of all the ways to mutate an ion channel, the number of ways that would result in its interacting with the base of some filament is surely in the distinct minority. And of all the ways to mutate an ion channel that gloms onto a filament, the number of ways to mutate it such that rotation does not occur is probably much higher than the number of ways to elicit some rotation. Thus, as with the first cooption event, we need another special mutation. This one allows some ion channel to glom onto the base of a filament and open its channel and expose the ion flow to the proto-rotor in such a way that a set of electrostatic interactions just happen to form and elicit significant rotation. Suffice it to say that such an improbable mutation has never been observed in nature or the lab. Of course, just because such a mutation may be highly improbable does not mean it did not occur. But if your thesis is built around the existence of improbable events, you need some type of independent evidence to support such claims. Especially as the whole problem of IC again raises its head. Still Held Captive by IC The appeal to cooption to explain the origin of rotation still fails to escape the grasp of IC. This is because the motor depends on three proteins: fliG, motA, and motB. Remove any one of these three proteins and you do not have a motor that is 2/3 as good as the whole. Remove any one of these three and you have a non-functioning motor. Consider fliG. The set of hydrophobic mutations above abolished only motility and not flagellation. Keep in mind that this set of mutations does not represent an exhaustive screen of all the residues crucial to fliG's torque generation (we'd have to add in other residues important for structure and those that play a role in torque generation). Yet they (or some similar class) are essential for motility. Yet without the motA/motB interactions, they have no known function. MotA/MotB, on the other hand, could plausibly exist as some ion channel prior to the existence of the flagella, but there is no evidence of this. And there is no reason to think that the residues crucial for motility (important for positioning and direct interactions) would be under any selective constraint prior to the supposed fortuitous interaction. Thus, as far as we can tell, all the information necessary for motor function provided no selective benefit until motility appeared. And we're left with the intuitive awareness that of all the various sequences that can be shared by any randomly chosen three proteins, the number of ways to hook up into some motor structure are far less likely than the ways that do not elicit a motor function. Of course, just because an interaction may be highly improbable does not mean it did not occur. But if your thesis is built around the existence of improbable events, you need some type of independent evidence to support such claims. Selective Motility? Another aspect of this motility component of the EFM hypothesis worthy of a critical look is the assumption that some kind of primitive, proto-motility function would be selectively advantageous. While a crude Darwinian "common sense" would seem to indicate this, I am not so sure. To appreciate why, we need to ask why it is that modern day bacteria move in a series of straight runs and tumbles. Why don't they simply swim straight for a food source instead of taking a convoluted path involving short bursts of straight runs interspersed with tumbles that randomly reorient them? In fact, bacteria will only be propelled by their flagella spinning about 100-300 times/sec for about 3-4 seconds. Why? We sometimes forget that the small-scale world of bacteria is much different from our macro-world. Bacteria are constantly being buffeted by water molecules and thus live in a "Brownian storm." The simple fact is that because bacteria are so small, they swim through a Brownian storm. Brownian motion will knock bacteria off course after 3-4 seconds. [4] And this highlights a serious problem with the EFM hypothesis. The flagellum is a highly sophisticated machine. Even if one believes it evolved, what we study today is the product of billions of years of evolutionary modification. Yet even this high sophisticated/highly evolved system barely overcomes the Brownian storm. Thus, just how advantageous would some proto-wiggle really be? Imagine a boat in the ocean during a tropical storm. Would a propeller that spun once every second really be any better than no propeller? In other words, it is possible that biologically significant motility on these scales depends on a minimal amount of system complexity and output that is out of reach in a Darwinian search beginning with simple states. To assure myself this was not the case, I did a PubMed search with the following search words: " partial motility flagella selective advantage" and it returned 0 hits. I obtained one hit with the search words partial motility selective advantage" and this was not a relevant study. Thus, this essential feature of the EFM hypothesis is without any evidential support. To sum this section up, let's consider more problems inherent in the EFM hypothesis The fortuitous interaction component of the story is incredibly vague and without any independent support and appears to be an ad hoc component invented merely to prop up an a priori belief that cooption was behind the origin of the flagellum. Flagellar motor function appears to require a high degree of specific interaction among the parts. The information content involved is likely to high. The fortuitous interactions invoked to elicit rotation appear to be highly improbable, in light of the likelihood that such functional interactions would represent a very small fraction of all possible types of interactions. Also, we're faced with positing special mutations again. The fortuitous interaction does not eliminate the problem of IC even at this microlevel. Invoking a selective benefit for some ill-defined "partial motility" is completely without evidential support. It would seem that some form of minimal motility function is required to cross the threshold of selective benefit. NEXT: In the next essay, I will consider the generic appeal to cooption in this example to determine if teleologists ought to abandon their thesis and adopt it instead. Citations. 1. The bacterial flagellar motor. 2. DeRosier, DJ. 1998. The Turn of the Screw: The Bacterial Flagellar Motor. Cell 93, 17-20. 3. Yamaguchi S, Fujita H, Ishihara A, Aizawa S, Macnab RM. 1986. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J Bacteriol 166(1):187-93. 4. Bacterial Flagella: Flagellar Motor. 5. Oplatka A. 1998. Do the bacterial flagellar motor and ATP synthase operate as water turbines? Biochem Biophys Res Commun. 249(3):573-8. 6. Jiadong Zhou, Scott A. Lloyd, and David F. Blair. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. PNAS, 95, 6436-6441 7. Lloyd, SA and Blair DF. 1997. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of E. coli. JMB. 266, 733744. ID THINK Evolving the Bacterial Flagellum Through Mutation and Cooption: Part IV Evolving the Bacterial Flagellum Through Mutation and Cooption: Part IV I have previously discussed the various evolutionary mechanisms proposed to overcome the problem posed by Irreducible Complexity (IC). [1] Evolution through the coincidental cooption of an alternative function (CCAF) remains the best non-teleological explanation for the origin of IC. Thus, it is not surprising that the EFM hypothesis builds around this type of thinking. But there are two ways to envision CCAF: simultaneous cooption or gradual cooption. For the sake of simplicity, let us pretend the flagellum is a six-part IC system, where A,B,C,D,E, and G are essential proteins and F* is rotary motility. A simplistic model of simultaneous cooption is shown in figure 1. Figure 1. Simultaneous CCAF Here A, B, C, D, E, and G all pre-existed the flagellum and had alternative functions in the cell. Then, through some type of fortuitous event, they all came together and spawned flagellar function. Generating a novel function through the chance conglomeration of six independent proteins seems highly unlikely for two basic reasons: 1. Cooption really needs the help of gene duplication in order to donate the components to the new system. Presumably A-E and G exist because they provide some other selective benefit to the organism. Thus, titrating off these components, and their functions, onto another system is likely to bring about deleterious effects in six other systems. Thus, to make this explanation more plausible, we'd have to invoke nearly simultaneous gene duplications in six independent spots in the genome and ensure they were expressed at the same time. 2. The flagellum is a machine, thus its components must physically interact in a stable fashion to carry out the series of coordinated movements that reverberate as a function of the initial energy input. A nice example of this can be seen with some FliN mutants that cause a complete loss in motility in bacteria that still had flagella. Such mutants led to the original interpretation that FliN played a direct role in the torque generation of the motor. But later work better explained these mutants as having reduced binding to their sites in the flagellum. [2] Thus, merely unstabilizing the interaction between FliN and its partners resulted in a complete loss of motility, highlighting the importance of these "well-matched" physical interactions. Since components A-E and G were all previously shaped by selection for various non-flagellar functions, it seems highly unlikely that they would just happen to have the "right" match of conformations to sufficiently interact and generate some proto-flagella. Thus, it is not surprising that evolutionary biologist, H. Allen Orr, dismisses this type of solution to the problem of IC: First it will do no good to suggest that all the required parts of some biochemical pathway popped up simultaneously by mutation. Although this "solution" yields a functioning system in one fell swoop, it's so hopelessly unlikely that no Darwinian takes it seriously. As Behe rightly says, we gain nothing by replacing a problem with a miracle. [3] Perhaps we should then turn to the more "Darwinian" version of CCAF, one that merely envisions gradual addition to the IC system while sustaining the components by giving them each an alternative function along the way. The evolutionary origin of the same sixpart system might come together gradually as in figure 2. Figure 2. The Gradual Cooptional Origin of the Flagellum. A-E and G are gene products. F is an unspecified function. F1-F10 represent 10 different flagellum-independent functions. F* is rotary motility. Here, we invoke 10 alternative non-flagellar functions to sustain individual parts and partial-IC systems. The advantage to this explanation is that it gets us as far away from simultaneous CCAF as possible. It captures the gradualism at the heart of Darwinism and helps to push each addition to the system closer to observable mutation effects, such as those seen in the generation of antibiotic resistance. That is, mutations in one gene at a time can add the component to the system. What's more, gene duplication can now more plausibly contribute to the account, as a duplication could have occurred prior to each cooption event. Yet Orr is still skeptical of this type of account: Second, we might think that some of the parts of an irreducibly complex system evolved step by step for some other purpose and were then recruited wholesale to a new function. But this is also unlikely. You may as well hope that half your car's transmission will suddenly help out in the airbag department. Such things might happen very, very rarely, but they surely do not offer a general solution to irreducible complexity. [3] His solution is to invoke Original Helping Activities [1, 3], which would make the scenario more complex as many cooptional events would simply aid and not yet be part of a truly IC structure (as defined by Behe). Mutations would later alter them and confer an essential status upon them. When viewed like this, IC ceases to pose any problem. Since IC is a function-dependent concept, and functions can disappear and emerge, IC states simply evolve. In fact, this viewpoint is so seductive that it has led many to believe a replay of life's tape (borrowing from Gould's metaphor) would simply generate something comparable to the bacterial flagellum. That is, while the actual flagellum we see may not exist in some replay, some comparable, complex motility structure would likely exist playing its role in filling the niche afforded by motility. Yet despite its seductive appeal, we must ask a simple question : does this explanation really account for the origin of the bacterial flagellum? We are not merely engaged in speculative philosophy here. We are talking about something that actually exists and has an actual history. Possible worlds are fun to think about, but historical claims must have more than an intuitive appeal to the possible to support them. History is not about what could have possibly happened; it is about what did happen. Thus, does this gradual CCAF explanation really explain the origin of something that exists, namely, the bacterial flagellum? That we can vaguely envision it happening means only that we should seriously consider it as an explanation. It does not mean we should adopt it as the explanation. It the previous three essays, I highlighted many of the problems with the EFM hypothesis. Let us then step back from this example to see if further problems exist. Reverse Engineering Julie Thomas originally surveyed the bacterial flagellum from the perspectives of Ur-IC and thematic IC. [4] Ur-IC is a postulated IC state that existed in the last common ancestral flagellum. Thematic IC focuses on the various functional roles entailed by the components of a machine to determine if the roles themselves exist in an IC relationship. Thematic IC might also be a helpful concept as we try to begin reverse engineering the flagellum. If we look to the standard E. coli flagellum, it is composed of eight subsystems with distinct functions: 1. The Base: Composed of FliF (the MS ring) and FliG(N-term), FliN, and FliM (the C-ring). These protein rings in the inner membrane are the first structures built. Removal of any of these genes results in the inability to further construct the flagellum, indicating they serve as the foundation of the flagellum. Also, as mentioned previously, the C-ring plays additional function roles. Only the Nterminal 200 amino acids of FliG seem important in these regards, as a mutant FliG with approximately 100 amino acids truncated off its C-terminus still forms flagella, but motility is lost. [2] 2. The Motor: Composed of FliG (C-terminal), MotA, and MotB. MotA/B play two roles as part of the motor: a) They serve as the stator against which the rotor moves and; b) They conduct protons (or sodium ions) that serve as the energy source for motility. The C-terminal domain of FliG directly interacts with the stator and plays an essential role in torque generation. 3. The Switch: Composed of FliN and FliM, along with other proteins that are part of the chemotaxis system. The switch allows the motor to change from a clockwise rotation to a counter-clockwise rotation (and visa versa). 4. The Export Machinery: Composed of flhB, fliQ, fliR, fliP, FliI, and flhA. These proteins form a core of the type III machinery discussed in Part I of this series. Also included, though much less conserved, are FliF, FliG, and FliN. This machinery exports proteins that will form the more distal components outside the inner membrane. 5. The Drive Shaft: Composed of flgB, flgC, fliE, and flgG. These proteins form a tube that crosses the periplasm and transmits the torque generated by the motor to the filament. 6. The Bushings: Composed of two rings, the L ring formed by flgH and the P ring formed by flgI. FlgG is the most distal driveshaft protein that probably interacts with these rings. These rings facilitate the driveshaft's penetration of the outer membrane. 7. The Hook: Composed of flgE. It transmits torque from the drive shaft to the filament. It is connected to the filament through hook-filament junction proteins, flgL and flgK. 8. The Filament: It acts as the "propeller." It is composed of flagellin (fliC) and the cap (fliD) discussed previously in Part II. We could theoretically eliminate many of the genes listed above if we reverse engineer our way to a simpler flagellar prototype. The base itself could possibly be composed of one protein. The motor could be composed of one membrane protein and use the base as the rotor. We can eliminate the switch, as this is more of luxury item. For example, when the motor is switched such than bacteria tumble, this merely amplifies the effects of Brownian motion to facilitate the reorientation of the cell. Simply turning off the motor could accomplish much of the same effect. In fact, this is how the flagella in Rhodobacter sphaeroides work. [5] The export machinery is hard to reverse engineer since it is still a black box. Nevertheless, let's be radical and assume it could function much the same with only three proteins. The drive-shaft could theoretically be composed of a polymer composed of only one protein. The L and P rings are not needed in gram-positive bacteria, since such bacteria lack outer cell membranes, thus we can eliminate this from the prototype (although it raises an interesting question about the origin of gram-positives and gram-negatives to be explored at a later date). Finally, it would seem you could eliminate the hook and filament proteins and simply have the drive shift polymer extend well past the outer membrane. This would leave us with the Base, the Motor, the Export Machinery, and a Filament, which is awfully similar to the assumption about original flagella inherent in the EFM hypothesis (where the filament and motor are added to the export machinery through CCAF). And this would leave us with a total of only six proteins. Of course, a six-protein IC system still poses an IC problem, but in light of the EFM hypothesis, it does not seem insurmountable. Yet the problem with the above approach is two-fold. Our ability to reverse engineer, in a realistic manner, is greatly hindered by our lack of understanding about the mechanistic interactions between the various components of the flagellum. Furthermore, our experience with designing nanotechnology is practically non-existent. Both factors suggest this six-part system may very well be over-simplistic and non-workable. Recall, for example, that the flagellum must be assembled and the act of assembly may place constraints on the minimal complexity of a rotary nano-propeller. For example, we have already seen in part II that filament assembly appears to be IC, requiring both flagellin and the cap. That is, the cap is not an added luxury item that merely seals off the filament (as was once thought). It is an essential assembly factor, playing the role of a processive chaperone. Thus, we can predict that further understanding of the flagellar components is likely to expand the list of essential players in the minimal flagellar prototype. The Ur-IC Flagellum Because of these limitations, it would be helpful to consider the flagellar components last shared by an ancestral flagellum. Thomas [4] originally did this by comparing the flagellar gene content of various distantly related bacteria. Allow me to do likewise and provide a list of flagellar genes found in Aquifex aeolicus, Bacillus subtilis, Escherichia coli, and Treponema pallidum. (Table I) Table I. Genes Likely Part of the Ur-IC Flagellum [6] Functional Role Gene Products Motor MotA, MotB, FliG (C-term) Base FliF, FliG (N-term), FliM/N Export Machinery FlhB, FliQ, FliR, FliP, FliI, FlhA Drive-shaft FlgB, FlgC, FlgG, FliE Hook and Adapters FlgE, FlgL, FlgK, FlgD Filament FliC, FliD Thus, 21 genes are shared by these four distantly related bacteria. Aquifex aeolicus are the most thermophilic bacteria, growing just below the boiling point of water. They are also thought to represent the earliest lineages to branch off the eubacterial tree. Bacillus subtilis is a gram-positive soil bacterium that can use a wide variety of carbon sources. Very similar bacteria (Clostridium) used to be thought most primitive. Escherichia coli represent the gram-negative proteobacteria and live in the digestive tracts of many organisms. Their flagella are among the most studied. Treponema pallidum is a spirochete whose flagellum is part of a rather specialized motility organelle known as the axial filament. As I mentioned, these four species are very distantly related, as seen by the phylogenetic tree constructed from 16s rRNA sequence (Fig 3). Furthermore, all four bacteria have experienced very different environmental pressures over the last several billions years. This strongly implies that these 21 genes were present in the last common ancestor of all eubacteria, thus comprising the Ur-IC flagellum. To further test this notion, I surveyed the flagellar genes of Thermotoga maritima since it is also a very deeply branching bacterium. According to the TIGR list of flagellar genes, everything in the Ur-IC list is represented, thus confirming what IC would predict.[7] Furthermore, this Ur-IC state has persisted for billions of years since it appeared. That billions of years of microbial evolution, in each lineage, have not imposed significant permutations on this IC core speaks to its true IC state. Figure 3. Eubacterial phylogenetic tree. Adapted from [8] Since the last detectable flagellum most likely contained these 21 genes (22, if we split FliG; 23 if we separate FliM and FliN), we can finally turn to the hypothesis of gradual CCAF to understand why it is so unconvincing. NEXT: How well do the EFM hypothesis and gradual CCAF explain the origin of the UrIC flagellum? Citations 1. TeleoLogic 7 2. Lloyd SA, Tang H, Wang X, Billings S, Blair DF. 1996. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J Bacteriol 178(1):223-31 3. H. Allen Orr 4. Julie Thomas 5. Shah DS, and Sockett RE. 1995. Analysis of the motA flagellar motor gene from Rhodobacter sphaeroides, a bacterium with a unidirectional, stop-start flagellum. Mol Microbiol, 17, 961-9. 6. I've split the FliG into its two separate functional domains. I've collapsed FliM/FliN into one species for three reasons reasons: a) Various studies have shown FliN and FliM to be closely related in a functional sense; b) In Bacillus subtilis, FliN is missing, but another gene, FliY, exists that appears to be a fusion of FliM and FliN (Mol Microbiol 1992 Sep;6(18):2715-23; and c) among all the groups compared, bacteria either had FliN, FliM, or both. For example, species lacking FliN had FliM. Species lacking FliM had FliN. 7. TIGR 8. Perry, JJ and Staley, JT. 1997. Microbiology: Dynamics&Diversity. Saunders College Publishing; Fort Worth. p. 405. Note: On 3/07/02, I changed the Ur-IC scoring from 19 to 21 genes. The original scoring did not include the rod protein FlgG because it was not listed in TIGR's flagellar genes for B. subtilis. Further analysis uncovered that in Bacillus, FlgG is flhP. FlgG has clearly been identified in B. subtilis. See Gene 1991 May 15;101(1):23-31. FlgD is listed as a component in E. coli, Thermotoga, and Trepenoma. Although apparently lacking in B. subitilis, it was present in B. halodurans. Using flgD sequence from B. halodurans, I BLASTed the B. subtilis and Aquifex genome, finding significant homologs. ID THINK Evolving the Bacterial Flagellum Through Mutation and Cooption: Part V Evolving the Bacterial Flagellum Through Mutation and Cooption: Part V In the previous essay (part IV), I attempted to present the non-teleological explanation for the origin of the bacterial flagellum, coincidental cooption of alternative functions (CCAF), in its strongest possible form. In figure 2, the evolution of the bacterial flagellum was nothing more than the gradual addition of parts over time. Yet there is one thing that is somewhat misleading about Figure 2. It only presents a picture of the evolution of a six-part IC system. But we've seen that the Ur-IC state of the bacterial flagellum likely entailed around 20 gene products. As a result, a figure outlining the gradual cooption of parts should contain at least 20 components (A-E, G-U), 38 preflagellar functional states (F1-F38), and 19 cooption events. In other words, to propose a series of IC systems that gradually increase in complexity, step-by-step, (a 3-part system becomes a 4-part system becomes a 5-part system, etc.), 38 non-flagellar functions are involved. One can attempt to cut down on the 38 functions by arbitrarily eliminating some (due to the ad hoc load of positing 38 unknown functions), but to do so, one must dip from simultaneous cooption (since we are constrained to explain the origin of these 20 functional sequences) and thus weaken the whole hypothesis [1]. When it is realized that the CCAF hypothesis is more involved than many think, various relevant insights will emerge. And the first important insight is that the EFM hypothesis fails to map to the gradual CCAF hypothesis. This is because the EFM hypothesis identifies only two cooption events, not nineteen. It also only attempts the identification of two preflagellar functions for flagellar components: a secreted protein (that somehow became the filament) and an ion channel (that somehow became the stator/motor). Two preflagellar functions is a long way from thirty-eight. If we are to take the EFM hypothesis seriously, we should consider it in light of what we know about the Ur-IC flagellum and map the three basic components of the story to the players that did in fact come into existence. As such, the export machinery would be represented by the N-terminus of FliG and FliM, FliN, FliF, FlhB, FliQ, FliR, FliP, FliI, and FlhA. The "filament" would be represented by everything from the drive-shaft to the cap, including FliD, FliC, FlgE, FlgL, FlgK, FlgB, FlgC, FlgG, and FliE. Finally, the motor would be represented by MotA, MotB, and the C-terminus of FliG. Thus, the EFM hypothesis, as formulated, can offer us nothing more than a two-step simultaneous cooption scenario as shown in Figure 1. Figure 1. The EFM Hypothesis. It begins with multi-component export machinery and invokes an initial cooption even to explain the origin of the filament. But because the "filament" of the flagellum is also a multi-component system, simultaneous, not gradual cooption is being invoked. Its non-flagellar function is not provided. The second cooption event, where an ion channel is added to create the flagellum, invokes the same thing. If we are to map the EFM hypothesis to the Ur-IC state, it fails as a gradual cooption scenario. To rescue the EFM hypothesis from the pit of simultaneous cooption (i.e., random assembly), one must begin to tease apart the yellow, blue, and pink boxes above and assign autonomous functional roles to each apart from the complex. Only such an effort will move us towards the gradual route of Figure 2 from the previous essay. Until such a successful effort is made, the EFM hypothesis remains a story about simultaneous cooption, where nine parts are being added to a nine-part machine, which finally accepts the addition of another three-part complex [2]. The IC Grip Not only does the EFM hypothesis fail to map to a gradual CCAF scenario, but it never truly escapes the grip of irreducible complexity. To appreciate this, let us envision the gradual formation of the flagellum in light of the EFM hypothesis and CCAF. Let us assume that the construction of the flagellum roughly reflects its evolutionary assembly through cooption (a molecular version of ontogeny reflecting phylogeny). After all, the EFM hypothesis adopts this strategy. The Rings When the flagellum is constructed, the first thing that is laid down is the M-ring (also called the MS-ring) composed of the FliF gene product [2]. After the M-ring is formed in the inner membrane, FliG is added. Finally, FliM and FliN are added, but they need the help of FliG. The result is the formation of the C-ring/switch complex/rotor. Yet here is the key point: "the switch complex forms prior to the other cytoplasmic substructures, including the export apparatus." [3] In other words, while the EFM begins with the export apparatus, in flagella, the switch must be formed first in order to form the export apparatus. Since the switch and rotor function only as part of the rotary flagellum, from the start the flagellum is constructed with its motility function "in mind." [4] At this point, we simply need to ask what function the M-ring served at the beginning? When one BLASTs through the bacterial, eukaryal, and archaeal genomes, FliF has no homolog anywhere. FliF does function as part of the type III system, but as explained earlier [5], it has significantly changed and is probably incapable of substituting for flagellar FliF. It likely remains as part of the type III system to facilitate the construction of the export apparatus and is not directly involved in export. The bottom line is that FliF not only has no homolog, but its function is flagellum-specific (if we ignore the more recently acquired role in type III secretion). Thus, the EFM hypothesis begins with a protein ring in the inner membrane that has no function. And the same basic story holds true for FliG, FliN, and FliM. These proteins have no flagellum-independent homologs and their function is flagellum-specific. And the FliFFliG/FliF-FliG-FliM/N complexes formed by hypothetical cooption events have no function; no function emerges as a consequence of turning a protein ring into a four-part system. But any non-teleological scenario must explain the origin of this four-part C-ring that anticipates the rotary flagella before we can even turn to the export apparatus. Recall the key feature to any cooption explanation is to provide a function to an IC component that is not dependent on the IC system. Thus, FliF, FliG, FliN, and FliM require alternative functions that predated the flagellum. Yet there is none. We can always assert the existence of some unknown functions in an ad hoc fashion, but if the only reason for this is to rationalize an a priori belief that the flagellum evolved, this would hardly be a convincing move. In fact, if the flagellum was indeed designed, we could nevertheless always imagine a CCAF explanation when asserting unknown functions in such an ad hoc manner (more on this later). Thus far, we are left with an M-ring without function. Then a four-part C-ring without function. Without functions for the independent parts and their progressive "intermediates," we are merely left with a veiled appeal to non-Darwinian random assembly. In other words, several unselected sequential steps are being proposed. Export The next step would be to add the export machinery. Recall this was the first step in the EFM hypothesis, highlighting how it never really got off the ground. Yet, the problem of IC also surfaces here. First, there are six conserved, independent type III components found in all flagella and type III secretory machines. This strongly suggests this is a sixpart IC subsystem. In fact, this hypothesis has been supported by a recent genetic analysis of loss of function mutants involving all six components of the type III system from Salmonella enterica [6]. When the mutants were analyzed one at a time, it was determined that all six components are required for export function. Thus, the CCAF hypothesis must come up with alternative functions for all of these six-components. And since there is no evidence that a subset of these six-components can carry out any biological function, it appears again that simultaneous cooption (i.e., random assembly) is again being tacitly postulated. Of course, we can again imagine unknown functional states for all six independent components, and their progressive partial assembly, but this is yet another ad hoc move. So far we have a 10-part assembly. Yet taking into consideration all the scientific data today, it would not be until the tenth part is added do we finally get a functioning "export apparatus." This means that one has been invoking non-Darwinian random assembly (unselected sequential additions) all along to explain the origin of almost one-half of the flagellum. The Driveshaft Finally, it's time to co-opt our filament. But now we're back to the problems discussed in Part II of this series [7]. If we count everything from the base of the rod to the cap, we're dealing with nine more proteins. We might be able to shrink the number by invoking gene duplication, but even this move would be debatable. Let's first consider FliE. A recent study has provided good evidence for the role and position of this component [8]. FliE appears to form a junction between the M-ring and FlgB, which is the proximal component of the driveshaft (or rod). Its primary role appears to be architectural. As the authors of this study note: The axial proteins form a long, continuous, hollow cylindrical structure consisting of the rod, hook, hookfilament junction proteins, filament, and filament cap. Not only is this structure important for the function of the flagellum as a motor organelle but its central channel or lumen is the physical pathway by which axial protein subunits reach their assembly destination, the tip of the growing flagellum. The component substructures are all built with the following common theme. The subunits lie on the so-called basic helix of a cylindrical lattice. This underlying local helical symmetry (not to be confused with the macroscopic helicity of the flagellar filament) means that, in principle, subunits could be added indefinitely like the steps of a helical staircase. This is in contrast to the substructures of the basal body such as the MS ring, which have closed annular symmetry and thus a fixed number of subunits (thought to be about 26 in the case of the MS ring protein, FliF). The rod and MS ring appear to abut each other closely. How do substructures with fundamentally different symmetries join together? A specialized zone might facilitate the junction. FliE seems a possible candidate for construction of such a junction zone....We propose that the primary role of FliE may be as a structural adapter between the annular symmetry of the MS ring and the helical symmetry of the rod and all subsequent axial structures. That FliE functions to bridge the different symmetries of the M-ring and driveshaft makes sense in light of the flagellum as a functioning whole, and its possible role in a partially completed proto-filament, incorporated by cooption, is completely obscure. It wouldn't even span the periplasm. It's similar to explaining the function of lug nuts without the wheels of a car. FliE is also an unusual protein with respect to the other proteins that form the rod and beyond. It is the only gene in its transcriptional unit and alpha-helices run throughout the protein (alpha-helices in the other rod proteins are restricted to the terminal ends). All in all, the hypothetical cooption of FliE, essential for forming the remainder of the rod and filament, doesn't make any sense. The interesting twist is that FliE should be the prime candidate for the "protein that fortuitously stuck" to the export apparatus as part of the EFM hypothesis. This is because it is the most proximal component of the rod, attached to the M-ring. Yet it does not form a filament. In fact, the authors in the study cited above note, "we propose that FliE not be called a rod protein, since it differs in so many ways from the rod and other axial proteins." And it gets more interesting. FliE is not part of the type III secretion machinery. This indicates that as part of the evolutionary transition from flagellum to type-III secretion, it was lost. This, in turn, indicates that FliE function is flagellum-specific. The export machinery works fine without it. It is only needed in light of the flagellum as a functioning whole. We now have an 11-part system that appears to have no functional advantage over the original 10 part "export system" strung together by random assembly. We can now begin to add the rod. FlgB is supposedly coopted and added to FliE and begins forming the "basic helix of a cylindrical lattice." But here things get tricky when trying to collapse the gram-negative bacteria with the gram-positive bacteria. The latter group merely possess a thick peptidoglycan cell wall outside the membrane, while the former has a more complex arrangement including a second outer membrane (the periplasm being the space between the two membranes). The problem faced with coopting a drive-shaft in a gramnegative bacterium is that a partial rod that does not span the periplasm and penetrate the cell wall and outer membrane provides no obvious utility. This seems to be thorny problem that will escape our analysis since the Ur-IC scoring assumes all eubacteria are related through a common ancestor and thus factors out the gram-negative-specific features. Regardless of the fuzziness that comes from treating gram-negative and gram-positive flagella as the same, let us consider the rod (driveshaft) composed of flgB, flgC, and flgG. Both flgB and flgC are small proteins with approximately 130 amino acids in all five of the distantly related bacteria used to score the Ur-IC state. FlgG, the most distal component of the rod, has an average size of about 260 amino acids in the same five species. An interesting feature of all three proteins, among all five species, is that no cysteine residues are found anywhere. Cysteine is one of the more rare amino acids found in proteins. Yet, if one surveys the codon usage in these five bacteria, we would expect 1.1% of the amino acids in an "average protein" to contain cysteine. There are 3156 amino acids among all 15 of these rod proteins (from the five species). Thus, we might expect to find 34 cysteines, but there are none. This is most interesting when we consider that all proteins have many amino acids that can mutate to cysteine with a single base pair substitution. In contrast, using the same codon usage data, the average amount of glycine residues we might expect to find is 217 (6.88%) and the 15 rod proteins actually contain 198 glycines. Thus, it would appear that the rod proteins are somewhat atypical as far as bacterial proteins go. The stoichiometries of these three proteins tell an interesting story [9]. Both flgB and flgC are present at about 6 copies per flagellum. FlgG is present at about 12 copies per flagellum. If the known helical symmetry of the filament, at 5.5 subunits per turn, extends into the rod (and this seems quite plausible), then the three components of the rod are ordered such that the most proximal one, flgB, forms one turn, followed by flgC and another turn, followed by flgG and two turns. Since a helical structure has no inherent constraint on assembly ("in principle, subunits could be added indefinitely like the steps of a helical staircase"), this suggests some factor extrinsic to these proteins is regulating this assembly in such an ordered fashion. And this raises the issue of IC. It would seem there is no reason why the rod should be built around three proteins instead of simply one. Yet these three gene products are found in all flagella, dating back to the putative ancestral flagellum. This suggests one protein is not sufficient to form a functioning flagellar rod. Furthermore, the size of these proteins among these five distantly related bacteria has been held relatively constant (Fig 2), despite billions of years of experiencing very different selective pressures. It would seem some form of constraint or specification is at work, as natural selection will not tolerate too much deviation. And these size constraints map back to the last common ancestral flagellum, indistinguishable from the first flagellum. Figure 2. Protein size among rod components in five distantly related bacteria. The rod proteins all share the following features: they are secreted by the type III export machinery; they all lack cysteine; they have maintained a relatively uniform size despite long periods of different selective pressures; they have no function apart from the flagellum; and they show an intriguingly ordered arrangement in forming the rod. In other words, if we are to find a cooptable part, and look in a way that is informed by the data, we need proteins that satisfy these specifications. Recall that the EFM hypothesis proposes that a previously secreted protein was coopted to become the filament. Since flgB is the most proximal component of the rod, it is the most likely candidate for the originally coopted part. To get a rough feel for whether or not these specifications would be satisfied by a typically secreted protein, I compared the size and cysteine content of flgB with 18 proteins secreted by the type III apparatus of Yersinia, Salmonella, and E. coli as listed by Hueck [10] (Figure 3). Figure 3. FlgB and proteins secreted by type III export machinery. Avg. FlgB size shown in black. Proteins containing cysteine in red. Since 8/18 virulence proteins contain cysteine, the exclusion of cysteine does not seem to be a prerequisite for export. The fact that flgB lacks cysteine may just be a common feature of small proteins, as no protein smaller than 200 amino acids contained cysteine. However, recall that flgG also lacks cysteine, although its average size is 260 amino acids, and secreted proteins ranging from 212-343 amino acids all had cysteine. In fact, the cysteine content among these five proteins is 0.92%, indicating they reflect the cysteine content of a typical bacterial protein. FlgG thus appears atypical in this respect. When we turn to protein size, it appears that flgB is atypical of secreted proteins. Its average size among the five distantly related bacteria is 131 amino acids +/- 6. Only one of the 18 proteins falls in this size range, yopJ, which induces apoptosis in cultured mouse macrophages. Thus far, we can see that although these 18 proteins shared in the ability to be exported by type III machinery, as a group they don't satisfy the specification for excluding cysteine, indicating that this specification is not a function of the export process itself. Neither is there any apparent bias towards protein size that would indicate flgB was coopted from a typical secreted protein. When this is coupled to the fact the flgB shows no homology to these 18 proteins, and that none of these secreted proteins form filaments (as far as I know), the hypothetical cooptable part that supposedly gave rise to flgB would appear to have had some rather flgB-specific properties. Gene Duplication A plausible explanation that could account for the rod proteins (flgB, flgC, and flgG) is gene duplication. That is, originally something like flgB may have been coopted and then expanded by gene duplication to give rise to these three gene products. This would have occurred prior to the last common ancestral flagellum, since all flagella have these three gene products. There are many lines of circumstantial evidence to support this hypothesis. These genes are found clustered together in the same operon. FlgB and FlgC are the same size, while flgG is essentially twice the size of either flgB or C (suggesting a gene fusion, followed by a duplication). And sequence similarities between all three have been previously proposed [11]. Yet despite all this evidence, the picture is rather ambiguous. The reason being that the similarities mentioned above may only reflect functional features rather than a historical origin. In fact, when researchers first sequenced these components, they expected similarities for these reasons alone: Some degree of similarity among the sequences of the axial proteins would not be surprising, for two reasons. The first is that, since the axial proteins together form a continuous filamentous structure, we might expect them to have a similar lattice and to share common structural elements that determine the lattice....The second reason for looking for sequence similarities among the axial components concerns the manner in which they are thought to be exported across the cell membrane. [12] Again, the similarities may simply be a consequence of the design and assembly of this type of rod-structure. While gene duplication is commonly used to explain the origin of IC (as in the case of the vertebrate blood clotting system), this may be a case when IC renders gene duplication an implausible explanation. Recall that flgB, flgC, and flgG are found in all bacterial flagella. Apparently, loss of one gene product cannot be compensated by the presence of the other structurally similar gene products. And recall the way they are laid down: six copies of flgB form one turn of the helical lattice, then six copies of flgC form the next turn, then twelve copies of flgG form two more turns. Is this crucial? Recall also that all subunits are needed to form a rod that can span the length of the periplasm. If a hypothetical ancestral rod was homogenous, being composed of only one gene product, it is difficult to envision the selectable nature of turning it into such a highly ordered, heterogenous structure. Without such a demonstration, there is no need to go beyond the default position that such similarities reflect functional constraints only. If we turn to the sequence similarities themselves, the functional hypothesis is strengthened. The rod proteins tend to be most conserved at the N-terminal and Cterminal ends. This makes sense in light of their assumed assembly, where the C-terminal end of one protein interacts with the N-terminal end of the next protein. We can imagine that the C-terminal end of flgB must interact with the N-terminal end of flgC to form the helical lattice (where the transition from flgB to flgC occurs). However, the C-terminal end of flgB must also be able to interact with the N-terminal end of an adjacent flgB to form the first turn of the lattice. Thus, it is not surprising from an engineering perspective that similarities in structure and sequence would be found. Nevertheless, the sequence similarities are not that convincing. First, I used E coli sequence to BLAST bacterial genes. When I searched with flgB sequence, 33 similar proteins with E values less than 10-4 were scored (anything with larger E-value is probably due to chance). All 33 were flgB homologs from other species. When I searched with sequence from E. coli flgC, 36 hits were uncovered, again all of them being flgC homologs. I then used ClustalX to align the sequences of flgB and then flgC from the five species representing the Ur-IC state. ClustalX will align and score with three categories: completely invariant positions (the same amino acid in the same position); conserved with "strong" amino acids (sequences with an amino acid at the same position that belongs to a class with very similar biochemical properties) and; conserved with "weak" amino acids (sequences with an amino acid at the same position with similar biochemical properties). Let's call these the Invariant Positions (IP), the Conserved with Strong Positions (CWSP), and the Conserved with Weak Positions(CWWP). Table I. Clustal Scoring with the first two flagellar rod proteins in Aquifex, Bacillus subtilis, E. coli, Thermotoga, and Trepenoma. Alignment IP CWSP CWWP flgB 11 14 14 flgC 19 20 14 flgB, flgC 2 4 7 It can be clearly seen from Table I that while flgB and flgC show decent sequence conservation (about 30% in flgB and 40% in flgC), it is mostly lost when the two genes are aligned together (down to 10%). In other words, there is decent sequence conservation within the gene groups that is essentially lost between the gene groups. This again supports the IC hypothesis, indicating that flgB and C function is not redundant and entails separate sets of information not found in some stem gene at the base of a gene duplication expansion. If we turn to flgG, its larger size requires more than a simple duplication account. A possible explanation that takes advantage of the fact that flgG is twice the size of flgB/flgC is shown in Figure 4. Figure 4. Hypothetical evolution of flgG from an flgB/C-like protein. We begin with an 130 amino acid flgB/C-like protein whose N-terminal and C-terminal ends are conserved. Then at step 1, gene duplication occurs and fuses the gene in tandem such that is now expressed as a 260 amino acid protein. During step 2, the conserved residues of the C-terminal end of the first copy and the N-terminal sequence of the second copy is lost by mutation, leaving a larger protein with conserved C-terminal and N-terminal ends necessary for lattice formation. The scenario outlined in Figure 4 would predict that the central region of flgG functions primarily as a spacer merely to create a larger protein. When one aligns flgB or flgC, most sequence similarity is found in the C-terminal and N-terminal 40 amino acids. Thus, we could roughly represent the sequence of both flgB and flgC (both being about 130 amino acids) as 40-40-40, where the red signifies the conserved regions. According to the scenario in figure 4, the original fusion would then look like 40-40-40-40-40-40 and then evolve into 40-40-40-40-40-40. If flgG arose in this fashion, we would expect to find the sequence conservation retained in the two ends and no significant homology in the central 160 amino acids. However, when the five flgG sequences are aligned and scored, 22 invariant and conserved positions are found in the C-terminal 40 amino acids, 19 are found in the N-terminal 40, and 51 are found in the middle 160 amino acids. While the ends are more strongly conserved, the middle region still shows 32% sequence identity, suggesting it is not simply function-less spacer. Next, I decided to look if these 51 conserved positions in flgG are explained by a fusion of flgB, flgC, or a fusion of both. There are four possible fusion products: the C-terminus of flgB fuses with the duplicated N-terminus of flgB, the C-terminus of flgC fuses with the duplicated N-terminus of flgC, the C-terminus of flgB fuses with the duplicated Nterminus of flgC, and the C-terminus of flgC fuses with the duplicated N-terminus of flgB. I attempted to align each possible fusion product with an alignment of the central 160 amino acids of flgG (flgG160). Nothing significant was found. Two alignments tried to fit the fusion between positions 20-110 of flgG160, one attempted to fit between regions 80-180 of flgG160, and the final one attempted to fit between regions 20-90 and 120-150. No alignment turned up any invariant positions or more than three "conserved" positions. Furthermore, around position 120 of flgG we find a consensus sequence of YTRDGSF. A search of flgB and flgC sequence with permutations of this sequence (YTR, TRD, RDG, DGS, GSF) failed to turn up any matches. To summarize what this means, the central region of flgG contains roughly 50 conserved residues when the five genes from very distantly related bacteria are aligned. That they have been conserved for so long suggests they are functionally important. Yet this information is not found in flgB, flgC, or their possible fusion. What about ends? They do appear similar to those of flgB and C. However, when either gene product is aligned against flgG, similarity scores are much worse compared that the alignments of the individual genes themselves (Table II). Table II. ClustalX alignment scoring. IP = invariant position; CWSP = conserved with "strong" amino acids; CWWP = conserved with "weak" amino acids. Residues in Cterminal and N-terminal 40 amino acids are counted only. Align IP CWSP CWWP FlgB 11 13 14 FlgC 17 15 9 FlgG 12 23 7 FlgG and FlgB 3 9 11 FlgG and FlgC 1 11 9 To conclude, while there are similarities between these three gene products, there is no need to posit gene duplication the account for them given a purely functional explanation is sufficient. NEXT: Continued analysis Citations 1. Recall that the advantage to gradual cooption is its simplicity, where only one protein has to match another rather than a group of proteins matching another group. 2. I am splitting FliG into its two functional domains. 3. Kubori, TK, Yamaguchi, S., and Aizawa, S-I. 1997. Assembly of the Switch Complex of Salmonella typhimurium Does Not Require Any Other Flagellar Proteins. J. Bacteriology 179: 813-817. 4. An interesting FliF mutation was isolated back in the 1980s (J Bacteriol 1989 Apr;171(4):2075-82). The mutant bacteria could swim fine in a liquid media, but upon placing it in a viscous medium, its basal body and filament dissociated from the M-ring. It's as if the torque generated by the motor ripped the flagellum in half when placed in a context that resisted rotation. This raises the interesting question of the importance of stable protein-protein interactions necessary for rotary motion. What are the minimal number of contacts needed to prevent the torque from twisting the flagellum apart and how do we place these without hindering flagellar assembly and rotation? 5. TeleoLogic 10 6. Anand Sukhan, Tomoko Kubori, James Wilson, and Jorge E. Galán. 2001. Genetic Analysis of Assembly of the Salmonella enterica Serovar Typhimurium Type III Secretion-Associated Needle Complex. J. Bacteriology 183: 1159-1167. 7. TeleoLogic 11. 8. Minamino T, Yamaguchi S, Macnab RM. 2000. Interaction between FliE and FlgB, a proximal rod component of the flagellar basal body of Salmonella. J Bacteriol 182:3029-36 9. Jones CJ, Macnab RM, Okino H, Aizawa S. 1990. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J Mol Biol 215:331 10. Hueck, CI. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Micro Mol Bio Rev 62: 379-433. 11. Homma, M, Kutsukake, K, Hasebe, M, Iino, T, and Macnab, R. 1990. FlgB, FlgC, FlgF, and FlgG. A family of related proteins in the flagellar basal body of Salmonella typhimurium. J. Mol. Biol. 211: 465-477. 12. Homma, M, DeRosier, DJ, and Macnab, RM. 1990. Flagellar hook and hookassociated proteins of Salmonella typhimurium and their relationship to other axial components of the flagellum. J. Mol. Biol. 213: 819-832. ID THINK http://www.idthink.net/biot/flag1/