Assiut university researches Genetical Studies on Peanut (Arachis
advertisement

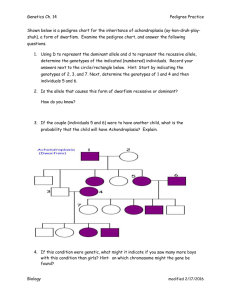
Assiut university researches Genetical Studies on Peanut (Arachis Hypogaea L.) Tissue Culture درا سات وراث ية ع لى زراعة االن سجة ف ى ال فول ال سودان ى Douha Salah Mohamed Mahmoud ضحى ص الح محمد محمود Adel Sayed Taghian, Hamdy Mohamed El-Aref, Bahaa El-Din ElSayed Abd-El-Fatah ب هاءال دي ن ال س يد ع بدال ف تاح، حمدى محمد ال عارف،عادل س يد ت غ يان Abstract: The present investigation was carried out at the Department of Genetics, Faculty of Agriculture, Assiut University, Assuit, Egypt, during 2009 – 2013. The investigation aimed at assessing the utility of peanut tissue culture for studying the following characteristics: I- Study the tissue culture response and plant regeneration in peanut (Arachis hypogaea L.). IIDetermine the genetic variability and polymorphism among six peanut varieties by RAPD, ISSR and SRAP markers, and to identify markers associated with tissue culture response. III- Assessing the possibility of obtaining salt tolerant peanut plants via tissue culture. VI- Studying the changes in gene expression under salinity stress as revealed by protein analysis. Six genotypes of peanut namely; Ismailia-1 (G1), Giza-5 (G2), Agryl (G3), Gregory (G4), Giza-7 (G5) and Giza6 (G6) were used in the present investigation. The results obtained could be summarized as follow: 1- Calluses and shoots were formed from the emberyonated and deemberyonated cotyledon explants of all genotypes on all tested media. The regeneration rate depended upon the genotype, type of explant and medium. 2- The MS medium supplemented with 25.0 mg/l BAP (SIM-P1 medium) was superior in callus formation (78.50%) and number of shoots per explant (10.28 shoots), which suggested that it was more suitable for regeneration of peanut plants than the other tested media. 3- The embryonated cotyledon was suitable explant for peanut tissue culture which exceeded the deembryonated one in callus formation, shoot regeneration and rooting of shoots. 4- The high efficiency of in vitro root formation could be obtained by culturing of regenerated shoots on MS medium supplemented with 1.0 mg/l NAA (Rzmedium). 5- The range of Euclidean distance among all genotypes using tissue culture traits was relatively wide (0.628 – 4.651), which indicated high amount of variation in tissue culture traits among the genotypes. 6- The UPGMA dendrogram based on tissue culture data separated the peanut genotypes into two clusters; the first one contained the highly responsive genotypes Giza-5, Gregory and Giza-7, while the second included the low responsive genotypes Ismailia-1, Agryl and Giza-6. 7- from the three molecular markers (RAPD, ISSR and SRAP primers), a total of 202 DNA fragments were amplified by 18 primers from all genotypes with an average 11.22 bands/primer. Out of these fragments, 71 (35.15%) showed polymorphism and 131 (64.85%) were monomorphic. 8- Four molecular markers generated by different primers [461bp (OPA06), 1480bp (HB09), 205bp (HB12) and 465bp (Em1c – me4)] can be considered as positive markers for tissue culture response in peanut. 9- RAPD, ISSR and SRAP markers showed positive correlation with tissue culture traits. 10- A total of 43 DNA bands were obtained from the six peanut genotypes using six RAPD primers and ranged in size from 772bp (OPA07) to 71bp (OPA01). 11- Only 37.21% of the DNA bands generated by RAPD were polymorphic while 62.79% were common between the tested genotypes. 12- Unique positive RAPD markers were detected for the genotypes Giza-6 (1 marker) and Giza-5 (2 markers) which could be used for genotype identification and discrimination. 13- Negative RAPD markers were detected for the genotypes Giza-7 (2 markers), Agryl (3 markers), Gregory (1 marker) and Giza-6 (1 marker). 14- A total of 60 DNA bands were obtained from the six peanut genotypes using ten ISSR primers and ranged in size from 1480bp (HB09) to 93bp (HB15). 15- Twenty-seven (45%) of ISSR bands were polymorphic while 33 bands (55%) were common between the tested genotypes. 16- Positive RAPD markers were detected for the genotypes Ismailia-1 (4 markers) and Giza-6 (1 marker) which could be used for genotype identification and discrimination. 17- A total of 99 DNA bands were obtained from the six peanut genotypes using six SRAP primer combinations and ranged in size from 1461bp (Em1a – me4) to 51bp [(Em1a – me2), (Em1c – me4) and (Em2 – me1b)]. 18- Twenty-eight (28.28%) of SRAP bands were polymorphic while the rest of bands (72.72%) were common between the tested genotypes. 19- One unique positive marker in Giza-6 and four negative markers in Gregory which were detected by SRAP primer combinations and could be used for genotype identification and discrimination. 20- Based on combined data of RAPD, ISSR and SRAP, the highest genetic similarity (0.951) and shortest genetic distance (0.049) were found between Giza-5 and Gregory. While the lowest genetic similarity (0.87) and longest genetic distance (0.130) was found between Gregory and Giza-7. 21- The dendrogram generated based on combined RAPD, ISSR and SRAP data separated the peanut genotypes into two clusters; the first one contained the highly responsive genotypes Giza-5, Gregory and Giza-7, while the second included the low responsive genotypes Ismailia-1, Agryl and Giza-6. 22- The development of callus and shoot differentiation were decreased with the increment of NaCl level from 50 to 150 mM. 23- At higher concentration of NaCl (200mM), the cultured explants turned brown and the few developed calli failed to regenerate shoots or regenerate abnormal shoots with very low frequency in most genotypes. 24- A total of 1760 embryonated cotyledons from Giza-5, Gregory, Giza-7 and Giza-6 were cultured under selection conditions of salinity stress (150 mM NaCl). 25- The regeneration rate was reduced under salinity stress to be 28.67% in callus and 15.15% in shoot formation of that on the control treatment. 26- The selected and unselected plants in comparison to their donor parents were tested for salinity tolerance in vitro on 0.0 and 150mM NaCl. The selected plants showed significant enhancement in their growth under salinity treatment, as compared with the unselected plants and their donor parents. 27- Salinity-stress induced the synthesis of two new protein bands (RF = 0.931 and 0.816) in all tested genotypes. While, one band at RF 0.978 in Giza-5 and other band at RF 0.532 in Giza-7 were also induced under salinity stress. 28- Salinity stress also reduced the expression of 5 proteins in G5, and 3 protein bands in G4 29Salinity stress also enhanced and/or decreased the expression of other proteins in the tested genotypes of peanut. 30- Salinity stress treatment lead to differential expression of the genetic information in peanut, resulting from changes in gene products, including induced/ enhanced the synthesis of certain proteins and simultaneously reduced/decreased the expression of other protein sets in the tested genotypes.