Identification of potential breast cancer treatment drugs
advertisement
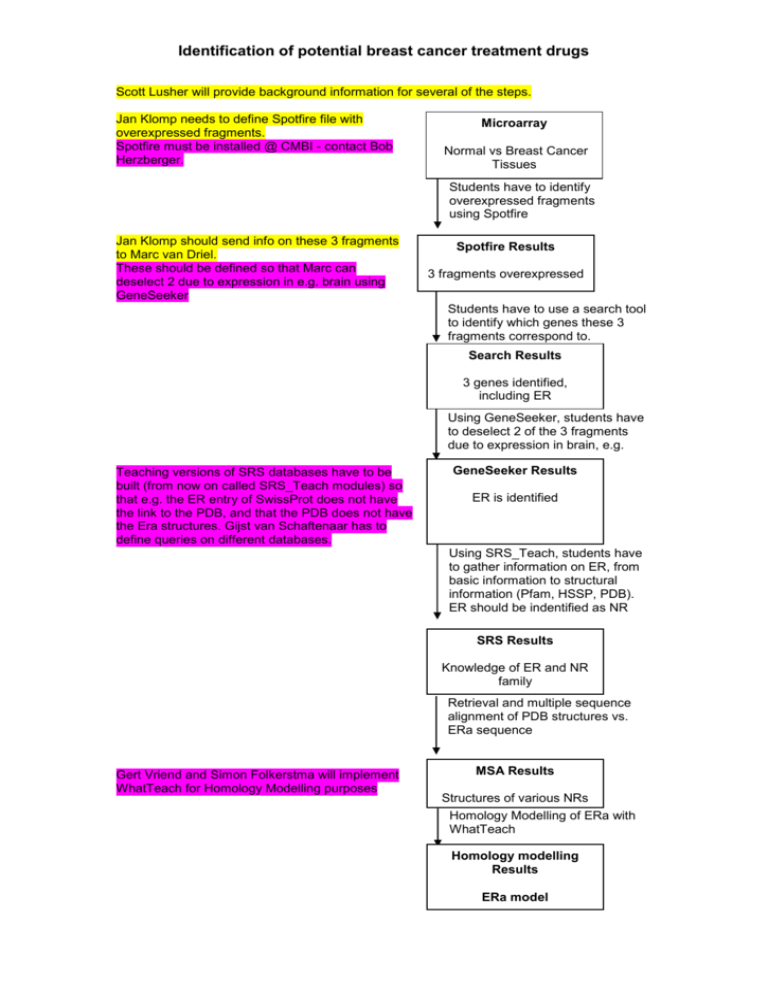
Identification of potential breast cancer treatment drugs Scott Lusher will provide background information for several of the steps. Jan Klomp needs to define Spotfire file with overexpressed fragments. Spotfire must be installed @ CMBI - contact Bob Herzberger. Microarray Normal vs Breast Cancer Tissues Students have to identify overexpressed fragments using Spotfire Jan Klomp should send info on these 3 fragments to Marc van Driel. These should be defined so that Marc can deselect 2 due to expression in e.g. brain using GeneSeeker Spotfire Results 3 fragments overexpressed Students have to use a search tool to identify which genes these 3 fragments correspond to. Search Results 3 genes identified, including ER Using GeneSeeker, students have to deselect 2 of the 3 fragments due to expression in brain, e.g. Teaching versions of SRS databases have to be built (from now on called SRS_Teach modules) so that e.g. the ER entry of SwissProt does not have the link to the PDB, and that the PDB does not have the Era structures. Gijst van Schaftenaar has to define queries on different databases. GeneSeeker Results ER is identified Using SRS_Teach, students have to gather information on ER, from basic information to structural information (Pfam, HSSP, PDB). ER should be indentified as NR SRS Results Knowledge of ER and NR family Retrieval and multiple sequence alignment of PDB structures vs. ERa sequence Gert Vriend and Simon Folkerstma will implement WhatTeach for Homology Modelling purposes MSA Results Structures of various NRs Homology Modelling of ERa with WhatTeach Homology modelling Results ERa model Identification of potential breast cancer treatment drugs Gert Vriend will implement a dedicated WhatCheck module ERa crystal structure is given to students Homology modelling Results ERa model Students have to compare model to crystal structure and evaluate quality of both using WhatCheck Gijst van Schaftenaar has to build a ca 20 compound mol2 database with active and non active compounds, including tamoxifen. Tamoxifen shoud be one of the best scoring compounds in the next docking step Basic understanding of ERa crystal structure FlexX docking of compound library. Comparison of docking poses. Students are provided with in-house compound library Paul Verwer has to decide which MD simulation program to use. MD shoud be run in advance and saved so that students have the results immediately. FlexX Results Selection of 2 compounds, including tamoxifen. Preparation of MD simulation runs for 2 selected compounds. Run of Whatcheck on optimised complexes MD Simulation Results Verification of improvement of quality of complexes. Identification of interacting residues within complex containing tamoxifen. Comparison to Simon's interacting residue database Interacting residues list Verification of lack of OH-histidine interaction in tamoxifen complex Proposals for modification of tamoxifen. Optimisation results Proposal of Dihydroxytamoxifen Docking/MD of dihydroxytamoxifen Final docking results Verification of improvement in interaction of Dihydroxytamoxifen with ERa










