File
advertisement

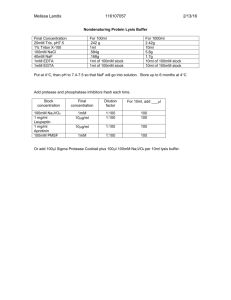
Immunoprecipitation (Adapted from eBioscience) Introduction Immunoprecipitation involves the interaction between a protein and its specific antibody, the separation of these immune complexes with Protein G or Protein A, and the subsequent analysis by SDS-PAGE. The protocol below offers a general guideline for immunoprecipitation for native protein at endogenous level. Optimization may be required for each specific antigen and antibody. The abundance of a given protein in a sample is variable and a critical factor for obtaining desired results The choice of lysis buffer is critical and dependent on the nature of the protein to be studied. NP-40, a non-ionic detergent, is the most commonly used detergent in cell lysis buffers. Increasing the salt concentration, decreasing the detergent concentration, or changing the detergent to Triton X-100, Saponin, Digitonin, CHAPS etc. are steps that can be taken to optimize conditions for immunoprecipitation. Table 1: Binding Characteristics of Some Immunoglobulins Binding Characteristics of Some Immunoglobulins Immunoglobulin Protein A Protein G Mouse IgG1 + ++ Mouse IgG2a +++ +++ Mouse IgG2b ++ ++ Mouse IgG3 + +++ Mouse IgM Mouse IgA Mouse IgE Rat IgG1 + + Rat IgG2a +++ Rat IgG2b ++ Rat IgG2c + ++ Human IgG1 +++ +++ Human IgG2 +++ +++ Human IgG3 +++ Human IgG4 +++ +++ Materials * Cell lysate or Tissue Lysate * Protein A or Protein G slurry * Primary antibody Buffers * Lysis buffer * Proteases Inhibitors (various brands, Roth turns out to be the cheapest and is effective) * PBS * SDS-PAGE sample buffer Instruments * Centrifuge * Rocking platform or rotator Method Step Ia: Cell Lysate Preparation 1. Harvest approximately 107 cells. Note: The total number of cells per ml and the cell equivalent loaded per lane of gel should be optimized specifically for each protein and antibody. 2. . Wash cells with ~10 ml of PBS in a conical tube and spin at 400xg for 10 minutes. 3. Discard supernatant and repeat step 2. Note: If using tissue culture dish adherent monolayer cells, wash cells in the flask two times with room temperature PBS and aspirate without disturbing the monolayer. 4. After the second wash, remove supernatant completely and resuspend the cell pellet in 1ml of cold Lysis Buffer containing 1X Protease Inhibitor Cocktail (final concentration of 107 cells/ml). Gently vortex the tube. Note: To prevent protease action effectively, the Lysis Buffer should be pre-chilled and all the following steps should be kept in the cold. The Protease Inhibitor Cocktail should be added to the required volume of the cold Lysis Buffer just before use. For adherent monolayers, add 1ml of cold Lysis Buffer containing 1X Protease Inhibitor Cocktail per 100mm culture dish. 5. Place the tube or the dish on ice for 30 minutes, with occasional mixing. 6. Spin cell lysate at 10,000xg for 15 minutes at 4°C. 7. Carefully collect supernatant, without disturbing the pellet and transfer to a clean tube. The cell lysate can be frozen at this point for long-term storage at minus 80°C. Discard pellet. Step Ia: Tissue Lysate Preparation 1. Harvest 2 cortices from 2 mouse brains or 6 hippocampi from 3 mouse brains 2. Add lysis buffer and triturate using mini-pestels, until the tissue is completely homogenized. 3. If there are still chunks of tissue, sonicate 3 x for 2” at medium frequency. 4. Read concentration we typically get a yield of 8-10mg of total protein. 5. Step II: Cell Lysate Preclearing 1. Transfer 50μl of the Magna_beads Protein G beads slurry (commercially available from several vendors, we have tried also Roth but this I would definitely not recommend!!!) to an eppendorf tube and add 450μl cold Lysis Buffer. Spin at 10,000xg for 30 seconds and remove the Lysis Buffer. Wash one more time with 500μl of cold Lysis Buffer. Resuspend the beads in 50 μl of cold Lysis Buffer. 2. Add this 50μl of prepared Protein G slurry and 500μl of Cell Lysate to an eppendorf tube and incubate on ice for 30-60 minutes. 3. Spin at shortly at 4°C and then transfer to the magnetic holder on ice, transfer the supernatant to a fresh eppendorf on ice. Step III: Immunoprecipitation 1. Add 5-10μg of antibody to the eppendorf tube containing the cold precleared lysate. Usually bad Abs do not work even when u put 50ul so make sure u have a good Ab to start with. 2. Incubate at 4°C for 1 hour. 3. Add 50μl of washed Protein G slurry in prechilled Lysis Buffer (prepared as instructed in Preclearing Step 1 above). 4. Incubate for 1 hour at 4°C on a rocking platform or a rotator. 5. Spin at shortly at 4°C and then transfer to the magnetic holder on ice. 6. Carefully remove supernatant completely and wash the beads 3-5 times with 500μl of Lysis Buffer. To minimize background, care should be given to remove the supernatant completely in these washes. After the last wash, aspirate supernatant and add 50μl of 1X Laemmli sample buffer to bead pellet. Vortex and heat to 90-100°C for 10 minutes. 7. Spin at 10,000xg for 2 minutes, then transfer to the magnetic holder, collect supernatant and load onto the gel. Supernatant samples can be collected and kept frozen at this point if the gel is to be run later. 8. Follow manufacturer’s instructions for SDS-PAGE. Stain the gel for visual analysis of the immunoprecipitated protein. Immunoprecipitation Buffers * NP-40 Cell Lysis Buffer: o 50mM Tris-HCl pH 8.0 o 150mM NaCl o 1% NP-40 * Protease Inhibitor Cocktail (100X): o PMSF, 5mg (50μg/ml) o Aprotinin, 100μg (1μg/ml) o Leupeptin, 100μg (1μg/ml) o Pepstatin, 100μg (1μg/ml) o 100% Ethanol bring up to 1ml, aliquot and keep at minus 20 Trouble shooting No binding Possible causes: Target protein is not present in sample? Make sure sample is appropriate Target protein lost or degraded? Prepare fresh lysate und add suitable protease inhibitors, keep sample cold at all times, avoid using frozen lysate Antibody not suitable for IP? Check datasheet, polyclonal antibodies generally perform better than monoclonal antibodies Wrong beads used? Check which beads (e.g. Protein A, Protein G) binds best to the species and subclass of primary antibody Antibody concentration too low? Increase concentration (check datasheet for recommended starting dilution) Antibody incubation time too short? Increase incubation time Protein A/G incubation time too short? Increase incubation time Interfering substances in sample? Make sure sample is free of BME, DTT and other reducing agents as they may destroy antibody function. Excessive detergents and extreme pH may interfere with antibody-antigen interaction Wash step too stringent? Decrease number of wash steps, reduce stringency (e.g. less detergent and salt or different detergent) Protein glycosylated? Glycosylation can mask the epitope by steric blocking preventing the antibody binding. Denature protein by heating up the lysate for several minutes High background/non-specific bands Possible causes: Non-specific binding to agarose beads or antibody? Perform a pre-clear step by incubating the lysate with Protein A/G/L agarose conjugate without the antibody Aggregated protein, membrane fractions etc. in lysate? Centrifugate lysate and remove aggregated protein before adding antibody Antibody concentration too high? Use a lower concentration Wash step to short or not stringent enough? Increase number of wash steps, increase stringency (e.g. higher concentration of detergent or salt) Contaminated buffers and equipment? Use freshly prepared buffers and clean equipment Dirty membrane? Do not touch membrane with bare hands Many specific bands Possible causes: Proteolytic degradation? Multiple bands below expected molecular weight may indicate proteolytic degradation of sample. Prepare new lysate and add fresh protease inhibitors 55kDa and 28kDa – heavy and light chain? Run sample under non-reducing conditions so IgG runs at 150 kDa