hw2
advertisement

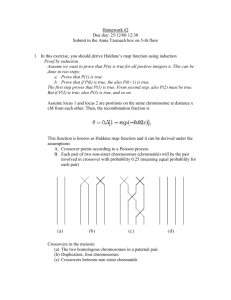
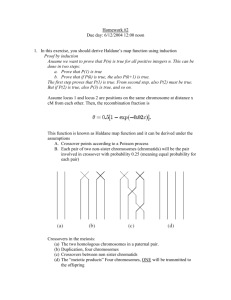
Homework #2 Due day: 25/12/06 12:30 Submit to the Anna Tzemach box on 5-th floor 1. In this exercise, you should derive Haldane’s map function using induction Proof by induction Asuume we want to prove that P(n) is true for all positive integers n. This can be done in two steps: a. Prove that P(1) is true b. Prove that if P(k) is true, the also P(k+1) is true. The first step proves that P(1) is true. From second step, also P(2) must be true. But if P(2) is true, also P(3) is true, and so on. Assume locus 1 and locus 2 are positions on the same chromosome at distance x cM from each other. Then, the recombination fraction is This function is known as Haldane map function and it can be derived under the assumptions A. Crossover points according to a Poisson process B. Each pair of two non-sister chromosomes (chromatids) will be the pair involved in crossover with probability 0.25 (meaning equal probability for each pair) Crossovers in the meiosis: (a) The two homologous chromosomes in a paternal pair. (b) Duplication, four chromosomes (c) Crossovers between non sister chromatids (d) The “meiotic products” Four chromosomes, ONE will be transmitted to the offspring *Homologous chromosomes: A pair of chromosomes containing the same gene sequences each derived from one parent. Notice, crossover counted iff happened between non-sisters chromosomes. Let N be the total number of crossover points. The Poisson assumptions implies P(n=k) –> K crossovers happened (1 Morgan corresponds to 1 crossover per chromatid , yielding the expectation 0.02x in the Poisson distribution). If N = 0, no crossovers have occurred and the probability that the chromosome transmitted to the offspring will be recombinant is 0. a) Prove (using induction) that the probability of recombination will be 0.5 for all N >=1 . Prove it for N = 1 (Hint: If only one crossover happened, then the result is .. recombinant and … non recombinant haplotypes..... Then assume for N = k that recombinant probability is 0.5 and prove for N = k +1 b) Prove, that it implies Haldane’s map function ( using θ = 0.5 * P(N>=1) ) 2. Does the “Genotype Elimination in Superlink” algorithm is complete. If yes, prove it. If no, show example. 3. Given next net Compute the most probable path for the sequence ACGT. Show your computations and pointers at each stage of the algorithm (you need not explicitly show terms that evaluate to 0). 4. Consider the following Bayesian network: Suppose that the net further records the following probabilities: Prob(A=T) = 0.3 Prob(B=T) = 0.6 Prob(C=T|A=T) = 0.8 Prob(C=T|A=F) = 0.4 Prob(D=T|A=T,B=T) = 0.7 Prob(D=T|A=T,B=F) = 0.8 Prob(D=T|A=F,B=T) = 0.1 Prob(D=T|A=F,B=F) = 0.2 a. b. c. d. e. Find Prob(D=T) Find Prob(D=F,C=T) Prob(A=T|C=T) Prob(A=T|D=F) Prob(A=T,D=T|B=F) 4. Consider next pedigree a. Is the pedigree consistent with the laws of Mendelian inheritance? b. Run Lange-Goradia algorithm c. Explain b. d. Suggest solution. Hint: look for O’Connell and Weeks “An optimal algorithm for automatic genotype elimination”