hw2
advertisement

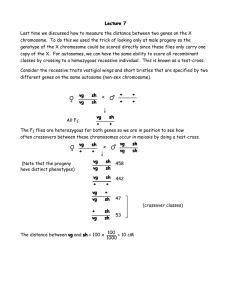
Homework #2 Due day: 6/12/2004 12:00 noon 1. In this exercise, you should derive Haldane’s map function using induction Proof by induction Asuume we want to prove that P(n) is true for all positive integers n. This can be done in two steps: a. Prove that P(1) is true b. Prove that if P(k) is true, the also P(k+1) is true. The first step proves that P(1) is true. From second step, also P(2) must be true. But if P(2) is true, also P(3) is true, and so on. Assume locus 1 and locus 2 are positions on the same chromosome at distance x cM from each other. Then, the recombination fraction is This function is known as Haldane map function and it can be derived under the assumptions A. Crossover points according to a Poisson process B. Each pair of two non-sister chromosomes (chromatids) will be the pair involved in crossover with probability 0.25 (meaning equal probability for each pair) Crossovers in the meiosis: (a) The two homologous chromosomes in a paternal pair. (b) Duplication, four chromosomes (c) Crossovers between non sister chromatids (d) The “meiotic products” Four chromosomes, ONE will be transmitted to the offspring *Homologous chromosomes: A pair of chromosomes containing the same gene sequences each derived from one parent. Notice, crossover counted iff happened between non-sisters chromosomes. Let N be the total number of crossover points. The Poisson assumptions implies P(n=k) –> K crossovers happened (1 Morgan corresponds to 1 crossover per chromatid , yielding the expectation 0.02x in the Poisson distribution). If N = 0, no crossovers have occurred and the probability that the chromosome transmitted to the offspring will be recombinant is 0. a) Prove (using induction) that the probability of recombination will be 0.5 for all N >=1 . Prove it for N = 1 (Hint: If only one crossover happened, then the result is .. recombinant and … non recombinant haplotypes..... Then assume for N = k that recombinant probability is 0.5 and prove for N = k +1 b) Prove, that it implies Haldane’s map function ( using θ = 0.5 * P(N>=1) ) 2. Maximum likelihood estimation. In the situations below, derive the maximum likelihood estimators of the parameters. Maximum likelihood estimation will be discussed in class. a. What is the maximum likelihood estimate of p when you record one observation from a Binomial(n,p) distribution? That is, X is distributed as Binomial(n,p). You observe X=x. What is your estimate of p? b. What is the maximum likelihood estimate of p when you record n independent observations from a Geometric(p) distribution? That is, X1, X2, ..., Xn are independent and distributed as Geometric(p) random variables. You observe X1=x1, X2=x2, ..., Xn=xn. What is your estimate of p? c. What is the maximum likelihood estimate of mu when you record n independent observations from a Normal(mu, 52) distribution? That is, X1, X2, ..., Xn are independent and distributed as Normal(mu, 25) random variables. You observe X1=x1, X2=x2, ..., Xn=xn. What is your estimate of mu? 3. Cystic fibrosis is a genetic disorder in homozygous recessives that causes death during the teenage years. If 4 in 10,000 newborn babies have the disease, what are the expected frequencies of the three genotypes in newborns, assuming the population is at Hardy- Weinberg equilibrium? Why is this assumption not strictly correct? 4. Below are the genotypic frequencies for two genes in a natural population. Answer the following questions. GENOTYPE A1A1B1B1 A1A1B1B2 A1A1B2B2 A1A2B1B1 A1A2B1B2 A1A2B2B2 A2A2B1B1 A2A2B1B2 A2A2B2B2 TOTAL FREQUENCY 13 92 173 22 164 313 9 73 141 1,000 a. Are the loci in Hardy-Weinberg equilibrium? b. The numbers of observed chromosome types are A1B1 = 304, A1B2 = 751, A2B1 = 113, A2B2 = 832 (assume a chromosome type is the same as a gamete type). Are these genes in linkage equilibrium? 5. Consider next pedigree a. Is the pedigree consistent with the laws of Mendelian inheritance? b. Run Lange-Goradia algorithm c. Explain b. d. Suggest solution. Hint: look for O’Connell and Weeks “An optimal algorithm for automatic genotype elimination”