Interpretation of Sequence Data
advertisement

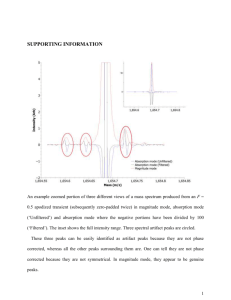
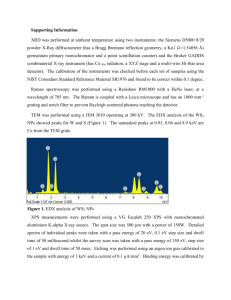
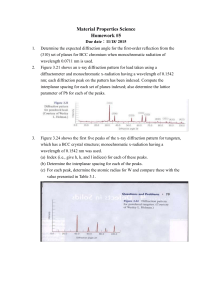
Interpretation of Sequence Data After the raw sequencing data has been collected by the 310 Genetic Analyzer and analyzed by the Sequencing Analysis program, it must be reviewed manually and interpreted with the aid of the Sequencher software. (See “Instructions for Sequencher 4.1”) It is important to remember that while software programs are very helpful in assigning base designations, every base call must be reviewed by human eyes and overridden if necessary. The computerized base calls are a guide, but the analyst has the final say. Guidelines for Interpretation Data consisting of distinct, relatively consistent peaks with little or no background noise is generally simple to interpret. Sometimes the data will present characteristics that make interpretation more complex, though not impossible. Interpretive skills improve with experience, but listed below are suggestions for those less familiar with mtDNA analysis on dealing with some of the more common features that affect interpretation. However, all data (especially indications of mixtures, heteroplasmy, etc.) should be reviewed on a case-by case basis, and interpretations made by the analyst based on his or her own expertise. The forensic community generally recognizes the HV1 region as consisting of base positions 16024-16365 and HV2 as base positions 73-340, as numbered according to the Cambridge Reference Sequence (also referred to as the Anderson Sequence). This laboratory will attempt to sequence all the bases within these defined regions, as well as an additional 25 base pairs in either direction (i.e. 15999-16390 and 48-365). While it is preferable to have confirmation of each sequence by comparing the forward and reverse strands, it is sometimes necessary to use two forward strands or two reverse strands as confirmation of the sequence. In these cases it is suggested that the sample be cycle sequenced a second time to obtain the confirmatory strand in the same direction. In addition to case samples, a positive and negative control must be sequenced. The purpose of controls is to show that each stage of the analysis is working properly. Therefore, only one positive and negative control is necessary for each step of the procedure (e.g. if samples from three amplification runs are combined into one cycle sequencing run, only one positive and one negative must be carried through to show the sequencing reagents and thermal cycler have performed as expected). If the results obtained for the controls are not as anticipated, the evaluation of the results will be determined on a case-by-case basis. Base Designations “A” designation—green peaks “G” designation—black peaks “T” designation—red peaks “C” designation—blue peaks “N” designation—peaks that, for whatever reason, are not clear enough to designate as A, G, T, or C. These bases are generally inconclusive. Often, a position that has an inconclusive (N) base in one direction may appear very clear in the other direction. Types of Polymorphisms Transitions: A G or C T (purines to purines OR pyrimidines to pyrimidines) Transversions: A C G T or T A C G (purines to pyrimidines OR pyrimidines to purines) Insertions: an extra base is present when compared to the Anderson reference sequence Deletions: a base is missing when compared to the Anderson reference sequence Interpretation of Results When mtDNA sequence data from a questioned sample is the same as that from a reference sample, the two samples cannot be excluded as originating from the same person or maternal lineage. When mtDNA sequence data from a questioned sample and a reference sample differ from each other by two or more differences, the samples are excluded as originating from the same person or maternal lineage (heteroplasmic differences will be evaluated on a case-by-case basis). When mtDNA sequence data from a questioned sample and a reference sample differ from each other by one difference only, the results will be evaluated on a case-by-case basis. The type of difference (e.g. heteroplasmy) and the relationship of the reference source to the evidence (e.g. a more distant maternal relative could be expected to have a greater number of sequence differences) will be considered. Data Interpretation In most cases, peaks in any given sequence will start out sharp and tall, then will slowly begin to fizzle out and become shorter and wider. This is a normal phenomenon that occurs as part of the nature of electrophoresis. Generally, the peaks will be of interpretive value for at least 250-400 base pairs, though the peaks at the end will not be as sharp relative to the peaks near the beginning. When the peaks deteriorate beyond an interpretable level as determined by the analyst, the remainder of the sequence should be cut off. Messy data at the beginning of a sample—it is common for the first 10-25 peaks to appear distorted while the rest of the peaks are quite normal. These initial peaks can be excluded from the region from which interpretations will be made (i.e. cut off). Messy data throughout a sample—these samples contain irregularities such as distorted peak shapes, inconsistent spacing, and background noise. The analyst’s own judgement will determine whether a sample is too messy to be of interpretive value. Low peaks—it is normal for peak heights in a sample to vary significantly, sometimes causing peaks to be very small (particularly C’s). Another common phenomenon is for a C peak to be lower than a “bridge” connecting T peaks that fall on either side of the C. Background noise—low peaks that appear over true sample peaks are referred to as background noise, and generally represent parts of the sequencing reaction that were not removed during purification (i.e. primers, unincorporated ddNTPs, etc.). Background peaks can range from only a few in the whole sequence to one or more under every true peak. The degree of background noise and the analyst’s expertise will determine the interpretive value of these samples. Irregular spacing—this may be evident in the form of occasional overlapping peaks or large gaps between peaks. Generally, this does not affect the quality of the data or its interpretive value. “Blown-out” peaks—large peaks that extend higher than can be viewed, generally occurring at the beginning (forward strand) or end (reverse strand) of a sample. It is sometimes possible for these peaks to be recognized individually and interpreted. Otherwise, simply cut off. Mixtures—the possibility of a mixture should be considered when more than one peak occurs at a particular base position that is not presumed to be due to background noise, messy data, irregular spacing, heteroplasmy, etc. A mixture of mtDNA from two people will show two different peaks at all of the base positions where one person’s sequence differs from the other. Interpretations regarding the sources of mixtures should be made very cautiously, if at all. Point heteroplasmy—occurs when at a particular base position, a person has one nucleotide in some cells and another nucleotide in other cells (e.g. in base position 16092 either a T or a C may be present). As a result, this base position will contain two peaks, provided each population of cells was amplified in large enough quantities to be detected. There is no limit as to how many positions in a particular sample may be heteroplasmic, but it is rarely seen occurring in more than one position in a particular sample. Length heteroplasmy—occurs when an extra base has been inserted at a particular position in some cells, but not in all cells, belonging to a particular individual. Length heteroplasmy is characterized by normal sequence data up to a particular base position, followed by peaks that appear “messy”. This is due to the fact that the peaks containing an inserted base are shifted by one base position and then superimposed over the peaks representative of the DNA that does not contain the insert. This is most commonly seen in the HV2 C-stretch region when an extra C has been inserted into some, but not all, of the sample DNA. When reviewing the reverse strand, the “messy” data will appear before the C-stretch, and the normal sequence data after the insert. C-stretch problem in HV1—A region exists in HV1 where a T is situated between two groups of C’s. When a transition polymorphism occurs changing the T to a C, the 310 Genetic Analyzer has difficulty reading through the lengthened “C-stretch”, and often the data beyond the C-stretch is of no interpretable value. When reviewing the reverse strand, the poor data will appear before the C-stretch, and the normal sequence data after it. Non-discrete peaks—these may occur when several of the same nucleotide appears in a row. For example, if the sequence includes the region ATGGGGGA, the G’s may be represented by one wavy peak as opposed to 5 distinct black peaks. Often the exact number of peaks present can be inferred from observing that region in the opposite strand. Dye blobs—usually occurring at the beginning of the sample, these are irregularly shaped “peaks” that actually represent residual ddNTPs. Dye blobs generally occur over top of the sample peaks without distorting the true DNA sequence. Spikes—spikes are generally due to particulate matter that has been picked up in the polymer and causes interference when the laser reads that part of the sample. A spike is usually very distinctive and appears as several irregular peaks occurring at one base position (sometimes encompassing two or three base positions). Peaks containing spikes may often be designated as N’s. Trailing/leading peaks—sometimes true peaks may have a much smaller peak of the same color preceding them or following them. When this phenomenon occurs, it is generally present throughout a significant portion of the sample and/or run, and does not greatly affect the interpretive value of the data. Negative samples / No DNA—chromatograms displaying peaks from which no useable sequence can be obtained may be due to an absence of DNA. These chromatograms generally have one or two predominant colors, and may include dye blobs, spikes, peaks near the baseline, many peaks on top of one another, or sections with virtually no peaks at all. The peaks observed on a negative sample result from unincorporated ddNTPs in the cycle sequencing mix, and will be present even in the absence of DNA.