tutorial
advertisement

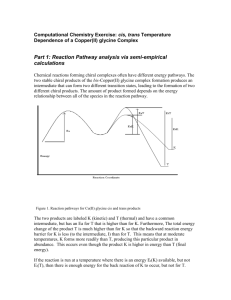
eQTL Explorer Demonstration Access to eQTL Explorer 1) Launch eQTL Explorer from http://web.bioinformatics.ic.ac.uk/eqtlexplorer/ 2) Login using eqtl_guest account. Username: eqtl_guest Group: public Genome display 1) Alter number of chromosomes in screen. Menu -> Options -> display -> 5 columns 2) Select both tissues and press fetch 3) How many eQTLs were retrieved for each tissue? Message at bottom of window Menu -> Summary 4) Notice what happens if you deselect trans or cis options. 5) Change C/T window value to 5Mb and press Fetch again. How many cis and trans eQTLs are retrieved this time? 6) Change C/T back to 10, change pvalue to 0.05 and press Fetch. Notice the relative increase in circles (trans eQTLs). Chromosome display 1) Click on chromosome 17 panel 2) Use sliding bar to zoom in and out of view 3) Mouse over eQTLs in large cluster near top of chromosome. What do you notice about the order of the eQTLs? 4) Click on eQTL. Follow probeset and marker links to external sources. 5) Mouse over pQTLs to get information about them. What is the difference between grey and white pQTLs? 6) Close chromosome panel Highlighting Probeset 1) Click on advanced options tab 2) In search by probeset field enter 1368526_at. Select “highlight probesets” from drop down menu 3) Return to main tab and press, ensure both tissues are selected and press fetch. 4) Notice there is no eQTL for this probeset in the kidney and the warnings tab reports this. 5) The cis eQTL in fat is ringed. NB The “fetch probesets” option will return only eQTLs that are formed by those probesets. pQTLs To identify cis eQTLs that co-localise with physiological QTLs 1) Go to advanced tab 2) Select “Body weight” and “Insulin Resistance” from pqtl options box. Click on checkbox “Display only these pqtls” 3) Set display options Menu -> Options -> Plot By: Probeset location 4) In main tab –select just the fat tissue 5) Uncheck trans and undefined display options, set pvalue to 0.01 and press Fetch. 6) Click on chromosome 3. Can you spot anything unexpected? Export 1) From chromosome view click on Table-> Show Table to get data for eqtls on that chromosome 2) From genome display. In main tab select tissue, pvalue, cis, tans options 3) In export tab. Scroll down and select ALL in chromosome box and press export. 4) Enter file name. Summary statistics 1 Experiment - BXH/HXB RI data genome v3.4 Genome Version - RGSC 3.4 P-value selected - 1.0E-4 LRS statistics = additive coefficient statistics = Cis/Trans window size - 10.0 Number of pQTLs - 251 --------------------------------------Redundancy Removal cutoff - 10 Analysis Id - 6 Tissue - fat Comment - No H26 Chip Name (source) - Rat Genome RAE230A Array (Affymetrix) Total number of eQTLs - 145 Total number of cis eQTLs - 126 Total number of trans eQTLs - 14 Total number of undefined eQTLs - 5 -----------------------Redundancy Removal cutoff - 10 Analysis Id - 7 Tissue - kidney Comment - No H26 Chip Name (source) - Rat Genome RAE230A Array (Affymetrix) Total number of eQTLs - 173 Total number of cis eQTLs - 149 Total number of trans eQTLs - 20 Total number of undefined eQTLs - 4 -----------------------Summary statistics 2 Experiment - BXH/HXB RI data genome v3.4 Genome Version - RGSC 3.4 P-value selected - 1.0E-4 LRS statistics = additive coefficient statistics = Cis/Trans window size - 5.0 Number of pQTLs - 251 --------------------------------------Redundancy Removal cutoff - 10 Analysis Id - 6 Tissue - fat Comment - No H26 Chip Name (source) - Rat Genome RAE230A Array (Affymetrix) Total number of eQTLs - 145 Total number of cis eQTLs - 118 Total number of trans eQTLs - 22 Total number of undefined eQTLs - 5 -----------------------Redundancy Removal cutoff - 10 Analysis Id - 7 Tissue - kidney Comment - No H26 Chip Name (source) - Rat Genome RAE230A Array (Affymetrix) Total number of eQTLs - 173 Total number of cis eQTLs - 137 Total number of trans eQTLs - 32 Total number of undefined eQTLs - 4 ------------------------