a. DNA Viruses - Lange Textbooks
advertisement

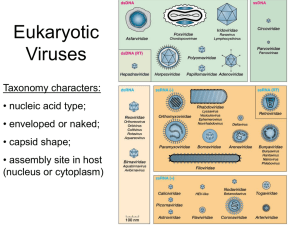
CHAPTER 6 The Nature of Viruses A virus is a set of genes, composed of either DNA or RNA, packaged in a proteincontaining coat. Some viruses also have an outer lipid bilayer membrane external to the coat called an envelope. The resulting complete virus particle is called a virion. They have an obligate requirement for intracellular growth and a heavy dependence on host cell structural and metabolic components. Therefore, viruses are also referred to as obligate intracellular parasites. Virus reproduction requires that a virus particle infect an appropriate host cell and program the cellular machinery to synthesize the viral components required for the assembly of new virions. The infected host cell may produce hundreds to hundreds of thousands of new virions, usually accompanied by cell death. Tissue damage as a result of cell death accounts for the pathology of many viral diseases in humans. In some circumstances, a virus fails to reproduce itself and instead enters a latent state (called lysogeny in the case of bacteriophages), from which there is the potential for reactivation at a later time VIRUS STRUCTURE 1. Viruses range in size from 20 to 300 nm in diameter 2. Naked capsid viruses have a nucleic acid genome within a protein shell 3. Enveloped viruses have a nucleocapsid (nucleic acid-protein complex) packaged into a lipoprotein envelope 4. Two basic shapes: cylindrical (helical) and spherical (icosahedral) 5. Outer shell is protective and aids in entry and packaging 6. Nucleic acid must be condensed during virion assembly I. GENOME STRUCTURE 1. The genomes of viruses can be either RNA or DNA, but not both 2. DNA or RNA genomes may be single or double stranded 3. Genomes may be linear or circular 4. Some genomes are segmented II. CAPSID STRUCTURE A. Subunit Structure of Capsids 1. The capsids or nucleocapsids are virus encoded specific proteins that protect the genome and confer shapes to viruses. 2. Capsids and nucleocapsids are composed of multiple copies of protein molecule(s) in crystalline array B. Cylindrical (Helical) Architecture 1. A cylindrical shape is the simplest structure for a capsid or a nucleocapsid. 2. Cylindrical viruses have capsid protein molecules arranged in a helix C. Spherical (Icosahedral) Architecture 1. Spherical viruses exhibit icosahedral symmetry 2. Capsomeres are surface structures composed of five or six protein molecules 3. Each structural subunit is referred to as a protomer. 4. To accommodate the larger cavity required by viruses with large genomes, the capsids contain many more protomers D. Special Surface Structures 1. Many viruses have structures that protrude from the surface of the virion 2. Surface structures are important in adsorption and penetration III. ENVELOPE STRUCTURE 1. Viral envelopes are lipid bilayer membranes containing virus encoded glycoproteins or spikes 2. Envelope glycoproteins, like surface proteins of naked capsid viruses, bind to the receptors on host cells for virus entry 3. Enveloped viruses are more sensitive to detergents, solvents, ethanol, ether and heat compared with viruses where outer coat is capsid protein. CLASSIFICATION OF VIRUSES 1. The classification of viruses considers various properties, including virions, genome, proteins, envelope, replication, physical and biological properties. 2. Virus families are designated with the suffix – viridae (like Herpesviridae), virus subfamilies with suffix virinae (Herpesvirinae), virus genera with suffix virus (Herpesvirus) 3. Virus species designated by a virus type (Herpes simplex virus I). General Features of Viral Growth 1. Virus Replication 1. Discrete phases: adsorption or attachment to the host cell, penetration or entry, uncoating to release the genome, synthetic or virion component production, assembly, release from the cell 2. Above describes what is called the productive or lytic response 3. Viruses can also produce a latent or nonproductive state in which no new virus is produced, the cell survives and divides, and the viral genetic material persists indefinitely 4. Some human viruses can cause oncogenic transformation 5. Viruses that can enter only into a productive relationship are called lytic or virulent viruses 6. Temperate viruses can either replicate or enter a latent state II. GROWTH AND ASSAY OF VIRUSES 1. Viruses are cultivated in cell lines or cell cultures derived from animal tissues 2. Cells taken from normal tissue cannot usually be propagated indefinitely 3. Cells taken from a tumor display long-term survival reflecting their tumor phenotype. 4. Permanent cell lines are useful for growing viruses 5. Cytopathic effects are characteristic for individual viruses 6. Viruses are quantitated by a plaque assay A. ONE-STEP GROWTH EXPERIMENT 1. One-step growth experiments are useful in the study of infections 2. Shortly after infection, a virus loses its identity (eclipse phase) 3. Infectious virus reappears at end of eclipse phase inside the cell 4. Proteins for replication are produced early and those for construction of virions are produced late Virus Replication Cycle I. ADSORPTION OR ATTACHMENT 1. A prerequisite is a collision between the virion and the cell 2. Adsorption involves attachment of viral surface proteins or spikes to the cell surface receptor proteins 3. Receptors for human viruses are usually glycoproteins located in the plasma membrane of the cell. 4. For some viruses like HIV coreceptors, are involved in adsorption 5. Viral spikes and phage tails carry attachment proteins 6. In some cases, a region of the capsid is the attachment protein 7. Adsorption is enhanced by presence of multiple attachment and receptor proteins 8. A virus is capable of infecting only a limited spectrum of cell types called its host range 9. Differences in host range and tissue tropism are due to presence or absence of receptors 10. Neutralizing antibodies are often specific for attachment proteins II. PENETRATION, ENTRY AND UNCOATING A. The Bacteriophage Strategy 1. Bacteriophage capsids are shed and only the viral genome enters the host cell 2. Tailed phages attach by tail fibers and DNA is injected through the tail B. Enveloped Human Viruses 1. Some enveloped viruses enter cells by direct fusion of plasma membrane and envelope 2. Protein spikes promote fusion of the viral membrane with the plasma membrane of the cell 3. Other enveloped and naked viruses are taken in by receptor-mediated endocytosis (viropexis) 4. Adsorbed virions become surrounded by the plasma membrane facilitated by the multiplicity of virion attachment proteins C. Naked Capsid Human Viruses 1. Naked capsid human viruses, such as poliovirus, enter the cell by viropexis 2. Acidified endosome releases nucleocapsid to cytoplasm 3. Virions may escape endosome by dissolution of the vesicles III. SYNTHETIC OR VIRION COMPONENT PRODUCTION 1. Important step in viral replication cycle because the virus must make mRNAs, proteins and genomes for the assembly of progeny or daughter viruses 2. Most RNA viruses replicate in cytoplasm, except influenza viruses and retroviruses that replicate in the nucleus 3. All DNA viruses replicate in the nucleus, except poxviruses that replicate in the cytoplasm IV. TRANSCRIPTION A. From Genome to mRNA 1. Virus-specified mRNAs direct synthesis of viral proteins 2. Most DNA viruses synthesize their mRNAs by using host RNA polymerase 3. Positive strand RNA virus genome serves as mRNA for early protein synthesis 4. Negative strand RNA viruses carry virion-associated RNA dependent RNA polymerase to produce initial mRNAs 5. There are a variety of pathways for synthesis of mRNA by different virus groups 6. Retroviral RNA is copied to DNA by virion reverse transcriptase; host RNA polymerase transcribes DNA into more RNA B. The Monocistronic mRNA Rule in Human Cells 1. For a viral mRNA to be recognized by the ribosome, its production must conform to the rules of structure that govern the synthesis of the host cell mRNAs 2. Prokaryotic (bacterial) mRNAs can be polycistronic which means it can contain the information for several proteins 3. Human virus mRNAs are almost always monocistronic 4. Most DNA viruses generate monocistronic mRNA through splicing 5. Some RNA viruses have segmented genomes to fulfill monocistronic mRNA rule 6. Some viruses (mainly negative sense RNA viruses) produce monocistronic RNAs by initiating synthesis at the start and pausing at the end of each gene 7. Positive sense RNA viruses (Picornaviruses, Flaviviruses) make a polyprotein that is proteolytically cleaved later into individual proteins C. GENOME REPLICATION a. DNA Viruses 1. In eukaryotic cells the enzymes and accessory proteins required for the replication of DNA are present only during the S phase of the cell cycle, and are restricted to the nucleus 2. The smallest DNA viruses depend exclusively on host DNA replication machinery 3. The largest DNA viruses code for enzymes important for DNA replication 4. Adenoviruses and herpesviruses code for their own DNA polymerases and other accessory proteins 5. Herpesvirus-encoded DNA polymerase is a target of antiviral therapy (e.g., acyclovir) 6. Viral processes that are distinct from normal cellular processes are potential targets for antiviral drugs 7. All DNA viruses except parvoviruses can transform host cells into cancer cells 8. All DNA polymerases synthesize DNA chains by the successive addition of nucleotides onto the 3′ end of the new DNA strand 9. They require a primer terminus containing a free 3′ -hydroxyl to initiate the synthesis 10. Replication of linear viral DNAs must solve the end problem b. RNA Viruses 1. RNA viruses mostly replicate in the cytoplasm and cells do not have RNA polymerases that can copy RNA 2. RNA viruses must encode their own polymerase or transcriptases 3. Transcription and replication must be separated for most RNA viruses 4. Picornaviruses use a protein to prime RNA synthesis D. ASSEMBLY OF NAKED CAPSID VIRUSES AND NUCLEOCAPSIDS 1. Enclosing the viral genome in a protein capsid is called assembly or encapsidation 2. Capsids and nucleocapsids self-assemble from preformed capsomeres a. Viruses with Helical Symmetry 1. Tobacco mosaic virus is a model for the construction of viral components 2. Doughnut-shaped disks containing a number of individual structural subunits are preformed and added stepwise to the growing structure. 3. The structural subunits as well as the RNA trace out a helical path in the final virus particle. 4. Features worked out for TMV probably apply in general to the assembly of the nucleocapsids of enveloped viruses. 5. For influenza and the other helical viruses with segmented genomes, the various genome segments are assembled into nucleocapsids independently and then brought together b. Viruses with Icosahedral or Cubic Symmetry 1. Icosahedral capsids are generally preassembled, and the genomes are threaded in 2. Phage heads, tails, and tail fibers are synthesized separately and then assembled 3. Some phage DNA is replicated to produce concatemers 4. Mechanisms for cutting phage DNA during packaging involve site-specific nucleases or headful cleavage 5. Host DNA may be incorporated by the headful mechanism, and generalized transduction results E. RELEASE a. Bacteriophages 1. Phages encode lysozyme or peptidases that lyse bacterial cell walls b. Human Viruses i. CELL DEATH 1. Naked capsid viruses lacking specific lysis mechanisms are released with cell death 2. Some viruses block or delay apoptosis to allow completion of the virus replication cycle ii. BUDDING 1. Most enveloped viruses acquire an envelope during release by budding 2. Poxviruses envelopes are wrapped from Golgi 3. The membrane site for budding first acquires virus-specified spikes and matrix protein 4. The budding process rarely causes cell death 5. Most retroviruses (except HIV) reproduce without cell death iii. CELL SURVIVAL 1. For retroviruses (except HIV-1 and other lentiviruses) virus reproduction and cell survival are compatible 2. Retroviruses convert their RNA genome into double-stranded DNA, which integrates into a host cell chromosome and is transcribed just like any other cellular gene 3. The virus buds through the plasma membrane without any permanent damage to the cell. 4. Filamentous phages assemble during extrusion without damaging cells QUANTITATION OF VIRUSES I. Hemagglutination Assay 1. For some human viruses, red blood cells contain receptors for the virion attachment proteins 2. An excess of virus particles coats the cells and causes them to aggregate and is called hemagglutination 3. Presence of hemagglutinin in the plasma membrane of the infected cell means that the cells will bind the red blood cells which is called hemadsorption 4. Hemagglutination can be used to estimate the titer of virus particles in a viruscontaining sample II. Plaque Assay 1. Plaque assay: dilutions of virus are added to excess cells immobilized in agar 2. Replicated virus infects only neighboring cells, producing countable plaques III. Immunological Assay 1. Viral antigen can be quantified by utilizing antigen-antibody specificity as measured by enzyme linked immunosorbent assay (ELISA) and immunofluorescence assays 2. Commercial antibodies detect or quantify the antigen of viruses in culture and body fluids, and 3. HIV can be quantified by the levels of an antigen (p24) in the culture fluid or blood IV. Molecular Assay 1. Viral genomes can be quantified to determine the amount of virus (viral load) in blood 2. The RNA genomes of the viruses are first reversely transcribed to cDNA and then amplified by polymerase chain reaction (PCR) 3. DNA genomes can be directly amplified by PCR to quantify the viral genomes Viral Genetics 1. Viruses generally utilize two mechanisms, mutation and recombination, by which viral genomes change during infection 2. Majority of the human virus particles from an infected cell are defective I. Mutation 1. Human viruses may code for their own DNA polymerases which are not as effective at proofreading as the cellular polymerases 2. Higher error rates in DNA replication endow the viruses with the potential for a high rate of evolution, but they are also partially responsible for the high frequency of defective viral particles 3. RNA viruses have even higher error rates because viral RNA polymerases do not possess any proofreading capabilities 4. RNA viruses commonly approach one mistake for every 2500 to 10,000 nucleotides polymerized 5. Because of the redundancy in the genetic code, some mutations are silent and are not reflected in changes at the protein level 6. Selective forces eliminate most mutants that fail to compete with the few very successful members of the population 7. With environment changes (e.g., neutralizing antibodies), a new subset of the population is selected and maintained as long as the selective forces remain constant 8. Changes lead to new variants not recognized by the immune system of previously infected individuals a phenomenon is called antigenic drift 9. High mutation rates permit adaptation to changed conditions 10. Mutations are responsible for antigenic drift in influenza viruses 11. High rates of mutation in retroviruses are due to error-prone reverse transcriptase 12. HIV-1 antigenic variation makes vaccine development difficult A. Von Magnus Phenomenon and Defective Interfering Particles 1. Defective interfering particles accumulate at high multiplicities of infection 2. Deletions result from mistakes in replication, recombination, or the dissociation–reassociation of replicases 3. Defective interfering particles compete with infectious particles for replication enzymes II. Recombination 1. Genetic recombination between related viruses is a major source of genomic variation 2. Bacterial cells as well as the nuclei of human cells contain the enzymes necessary for homologous recombination of DNA 3. Homologous recombination is common in DNA viruses 4. Recombination for viruses with segmented RNA genomes involves reassortment of segments 5. Segment reassortment in mixed infections probably accounts for antigenic shifts in influenza virus 6. Poliovirus replicase switches templates to generate recombinants 7. The diploid nature of retroviruses permits template switching and recombination during DNA synthesis III. THE LATENT STATE 1. The latent state involves infection of a cell with little or no virus production 2. Latent virus may be silent, change cell phenotype, or be induced to enter the lytic cycle IV. LYSOGENY 1. E. coli phage λ may be lytic or latent 2. When λ is integrated, the only active gene encodes a repressor for the other phage genes 3. Inactivation of repressor causes induction and virus production 4. Latent genomes can exist extrachromosomally or can be integrated 5. Phage λ integrates by site-specific recombination 6. Excision after λ induction involves recombination at junctions between host DNA and prophage 7. Specialized transduction occurs because excision occasionally includes genes adjacent to the phage genome 8. Lysogenic conversion results from expression of a prophage gene that alters cell phenotype 9. Several bacterial exotoxins are encoded in temperate phages