Paper
advertisement

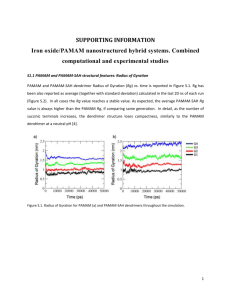
COMPUTER SIMULATION OF POLYAMIDOAMINE DENDRIMERS AND THEIR COMPLEXES WITH CISPLATIN MOLECULES IN WATER ENVIRONMENT N.K.Balabaev1, V.V.Bessonov1, I.M.Neelov2,3, M.A.Mazo4 1 Institute of Mathematical Problems of Biology RAS, Pushchino, 142290 Russia 2 School of Physics and Astronomy, University of Leeds, Leeds, LS2 9JT United Kingdom 3 Laboratory of Polymer Chemistry, University of Helsinki, P.O.Box 55, Helsinki, Finland 4 Institute of Chemical Physics RAS, Moscow, 119991 Russia ABSTRACT. Molecular dynamics simulations with explicit water were carried out for guest-host systems on the base of PAMAM-4.5 dendrimers and cisplatin PtCl2(NH3)2 molecules. Single dendrimer molecule and cisplatin molecules chemically attached to dendrimer terminal groups or adsorbed on the macromolecular surface were considered. AMBER force field, TIP3P water molecules and periodical boundary conditions were used for calculations. It is no protonated amines of PAMAM that correspond to pH 10. Computer experiments were conducted at temperatures 293, 310, and 350 K and pressure 1 bar. The structure and dynamics of guest-host systems was analysing. In all considered cases the dendrimers form a compact globule, which shape is far from spherical. Moreover the dendrimer cores dispose on the molecule surface in all considered cases. The chemically attached cisplatin penetrate into dendrimer deeper then non-attached one and decrease a large-scale intramolecular mobility. 1. Introduction The compound cis-PtCl2(NH3)2 (cisplatin), has become one of the most widely used drugs for the treatment of cancer [1,2]. Its mechanism of interaction with cells has been studied on a molecular level and it is well established, that death of the cell is induced by complexes of the platinum compound to two adjacent guanine bases [2]. But its remarkable anticancer properties can be accompanied by marked toxic effects as well as the development of resistance to the drug. Recently a polyamidoamine (PAMAM) dendrimer generation 4.5 was conjugated to cisplatin giving a dendrimer-platinate (dendrimer-Pt) which was highly water soluble and released platinum slowly in vitro [3,4]. In these works was shown that the dendrimerPt improved cisplatin efficiency and was less toxic (3- to 15-fold). PAMAM dendrimers are the polymers with unidispersed and well-defined molecular structures. These molecules can be synthesized in large quantities and have a large number of potential biomedical applications [5,6]. Highly branched, functionalized polymers have potential to act as a gene delivery and as efficient drug carrier systems [5,7-9]. There are a lot of publications devoted to the study of structure and mobility both single dendrimers, and dendrimer-guest systems. However till now our knowledge of molecular structure of this systems are rather fragmentary. The essential contribution to our understanding of the dendrimer molecules structure was achieved by molecular dynamic (MD) simulation [10-13]. Recently, more exhaustive atomistic MD simulations of dendrimers were carried out [14-25] including simulation PAMAM molecules [17,19,21,24] and research a solution be4 - 24 havior of dendrimers in explicit solvent [20,22,24]. Unlike simple coarse-grain dendrimer models the simulation of detailed molecular systems is rather complicated, as requires a substantiation for the large number of used parameters, very expensive, the received results, as a rule, and do not suppose wide generalization. However now it is the only way to receive the detailed information on spatial structure and mobility of separate molecules in solvent. In this study, we used atomistic MD simulations with explicit water to study the guest-host systems on the base of PAMAM-4.5 and cisplatin molecules chemically attached to dendrimer terminal groups or adsorbed on the macromolecular surface were considered. 2. The model and simulation details Calculations were performed for three systems: a) water solutions of PAMAM4.5, b) water solutions of PAMAM-4.5 with cisplatine, and c) water solutions of dendrimer-Pt molecule. PAMAM have tetrafunctional core –NCH2CH2N– with three radiating branches of –CH2CH2CONHCH2CH2N– and 64 terminal groups –CH2CH2COOH (Fig.1a). The dendrimer–Pt made up by joining 7 cisplatines –PtCl(NH3)2 to random choosen termal groups (Fig.1b). The molecular weights of cisplatine, PAMAM-4.5 and dendrimer-Pt are correspondingly 299 a.u., 11380 a.u. and 13164 a.u.. The calculation cell contained one dendrimer molecule, explicit water molecules, and 8 cisplatin molecules in system b (Tabl.1). OH OH OH O O OH OH O O O O OH O OH N O OH O N N O O N OH O N O OH O OH N O O N O N N O N N N N O OH OH O N N O NH3 NH3 O N N Pt O OH O Cl O N N O OH O OH OH O OH OH N OH O OH O O O O OH OH OH O O O O O O OH OH O NH3 O OH O OH O OH NH3 Cl Cl NH3 OH O Pt NH3 Pt O OH O OH O OH a b Fig. 1. Schematic drawing of PAMAM-4.5 (a) and dendrimer-Pt molecule (b). Tabl. 1. Some details of considered systems. Number Density System of water at 293 K, molecules g/cm3 a) PAMAM-4.5 3230 1.05 b) PAMAM-4.5 + cisplatin 3274 1.07 c) Dendrimer-Pt 3230 1.07 a) for –PtCl(NH3)2. 4 - 25 Dendrimer weight concentration 16.0% 18.6% 18.5% Cisplatin weight concentration 3.9% 2.6%a) The AMBER force field [26] was used for calculation. The potential energy comprising potential terms of bond Ub, angle Ua, torsion Ut, van der Waals Uvdw and electrostatic Ue interactions was used: U b K l (l l0 ) 2 , U a K ( 0 ) 2 , U t K [1 cos(n0 )], U vdw U LJ (rij )W (rij ) , where U LJ 4 ij [( ij / rij )12 ( ij / rij ) 6 ] and W(rij) is the switching function in interval 0.9 rij 1.05 nm, U e [qi q j /( rij )]We (rij ), where We is the screening function (Re = 1.05 nm), (1 rij / Re ) 2 , rij Re We (rij ) 0, rij Re In this equations the following notations are used: l is the bond length, is the bond angle, is the torsion angle, l0, 0 are equilibrium values for the bond lengths and angles; Kl , K, K are force constants for the bonds, angles, dihedrals angles, respectively; n0 is the dihedral multiplicity; rij is the distance between nonbonded atoms i and j; ij , ij are Lennard-Jones parameters for the atom pairs, qi , qj are the partial charges on atoms i, j, is the dielectric constant, Re is the screening radius. It is no protonated amines of PAMAM that correspond to pH 10. The H2O geometry parameters, the partial charges [27], and the force constants [28] for TIP3P water molecules are fixed. The parameters of force field and partial charges of cisplatin were taken same, as in [29]. Periodical boundary conditions were applied and the cells size was large enough (4.83 nm) to exclude any intaraction between dendrimers. Molecular dynamics techniques [30] were used for the equilibration and regular simulation. Collisional thermostat [31] and Berendsen barostat [32] were used for temperature and pressure support. The integration time step t=0.5 fs was used and the times of regular runs were 1 ns. Initial structure of a complex polymer system (coordinates and velocities of all atoms) plays the key role for successful modeling of its behavior. The preparation of representative structure of the system is usually complex and expensive procedure. Some technology was elaborated to construct dendrimers under consideration. At the first stage the special procedure was used to assemble the dendrimer structure, which was like a dandelion flower (Fig.2). A combination of constructor and collisional dy- 4 - 26 a b c Figure 2. Simulated snapshot of initial dandelion structure at the first stage (left) and configurations of dendrimers after 1 nsec runs (right). (a) PAMAM-4.5, (b) PAMAM-4.5 and cisplatin, (c) dendrimer-Pt. Water molecules are not shown to not complicate a picture. 4 - 27 namics computer programs were used to build up each next generation of isolated dendrimer molecule during this procedure. At the second stage collissional molecular dynamics technique was applied to equilibrate initial configuration of the dendrimer molecule. Than the macromolecule was immersed in water and equilibration of the total system was accomplished. The structure received was initial one for productive run of the system. 3. Results and discussions One of the characteristics of the dendrimer size is the radius of gyration RG. Its values averaged over the whole trajectory are given in Table 2. From this table we notice that this value of PAMAM in water correlate well with the evidence of another authors [14,18,20]. The radiuses of gyration of dendrimers with adsorbed and with chemically attached cisplatin are bigger and at the same. That curiously enough so in the former case the cisplatin molecules were not taken into account along calcuculations. In all considered cases the dendrimers forms rather compact globule, which shape is far from spherical (Fig. 2). This is apparent also from Table 2 where the values of the main radiuses of inertia R J / M (J are the principal moments of gyration tensor, =1,2,3, J1> J2 > J3) and relative values J2/J1 and J3/J1 are shown. The difference of this ratios from 1 characterise the deviation of dendrimer shape from the sphericity. In our case we see a strong asymmetrical molecules that consistent with the another simulation data for PAMAM [14,18,20], and the deviation from the sphericity increase in the presence of cisplatin. The dendrimer size and shape are independent of temperature at considered interval of temperatures. Tabl. 2. The radiuses of gyration RG (nm), the main radiuses of inertia (nm), and the relative values J2/J1 and J3/J1 for PAMAM-3.5 and dendrimer-Pt molecules. PAMAM-4.5 PAMAM-4.5 Dendrimer-Pt in water solvent in water solvent in water solvent with cisplatin 293 K 310 K 350 K 293 K 310 K 293 K 310 K RG 1.33 1.34 1.34 1.45 1.44 1.47 1.48 R1 1.22 1.22 1.23 1.36 1.36 1.40 1.40 R 1.11 1.11 1.11 1.20 1.21 1.22 1.21 R 0.89 0.93 0.93 0.94 0.92 0.95 0.96 J2 / J 1 0.84 0.82 0.82 0.78 0.79 0.77 0.78 J3 / J 1 0.54 0.55 0.57 0.47 0.46 0.46 0.46 The internal structure of the dendrimer and the distribution of the solvent inside of it can be seen from radial density distribution functions for dendrimer and solvent atoms relative to the centre of mass (CM) of the macromolecule (Fig. 3a). By calculation of the density distribution, all atoms were treated as uniform spheres with corresponding van-der-Waals diameters. It is seen that water molecules are not incorporated into the dendrimer. Only one water molecule dispose near the dendrimer CM in the system with adsorbed cisplatin. The density profiles are not essentially changed with the temperature. It was rather unexpectedly to discover that the dendrimer core during the run was far from CM the center of mass of the macromolecules and for PAMAM-4.5 in water even is farther, than for another cases (Fig. 3b). Moreover as can be seen in Fig.4 the dendrimer cores dispose on the molecule surface. So asymmetrical structure 4 - 28 is likely to be characteristic for small generation of PAMAM in water at high pH, that distinguishes its from carbosilane and polyamindoamine dendrimers [18,24,33]. g /c m 3 g /c m 3 G0 0 ,0 6 2 ,5 1 2 3 0 ,0 5 1 2 3 2 ,0 D e n d r im e r 0 ,0 4 1 ,5 0 ,0 3 W a te r 1 ,0 0 ,5 0 ,0 0 ,0 0 ,0 2 0 ,0 1 0 ,5 1 ,0 1 ,5 R, nm 2 ,0 0 ,0 0 0 ,0 0 ,2 0 ,4 0 ,6 0 ,8 1 ,0 R, nm 1 ,2 1 ,4 a b Fig. 3. Radial density distribution relative to the centre of mass of dendrimer at T=293 K. (a) Dendrimer and water atoms; (b) the contribution of dendrimer core atoms. 1 – PAMAM-4.5 in water solvent; 2 – PAMAM-4.5 in water solvent with cisplatin; 3 – dendrimer-Pt in water solvent. a b Fig. 4. Snapshots of PAMAM with adsorbed (a) and chemically attached (b) cisplatin at 293 K. Here the dendrimers are shown as a ball-stick model while the core and cisplatin as a spacefilling union of spheres model where each atom is drawn as a sphere of its Van der Waals radius. The allocation of the adsorbed and chemically attached cisplatin in dendrimer is considerably differing: in the latter case it penetrates deeper in macromolecule (Fig. 5). Chemically non-connected cisplatin can desorbs as may be seen on snapshots (see, for example, in Fig.2b) and as evidenced a high probability to find out the cisplatin on distances more than 2.3 nm from CM (Fig.5). 4 - 29 g/cm3 Cysplatin 0,4 Fig. 5. Radial density distribution of cisplatin relative to the centre of mass of dendrimer at T=293 K. 1 – adsorbed cisplatin; 2 – chemically attached cisplatin. 0,3 1 2 0,2 0,1 0,0 0,0 0,5 1,0 1,5 R, nm 2,0 2,5 3,0 Intramolecular dynamics of dendrimers was assessed as a mobility of branch centers and hydroxyl hydrogen atoms of the end groups. For that we calculated the time dependences of the distances L(t) between concerned atoms and CM. As would be expected the atom mobility depend on temperature, but even at temperature 293 K L(t) of some ends groups vary more than 0.8 nm during 1 ns for all cases. However the mobility of chemically connected and adsorbed cisplatin is essentially different (Fig.6). In the first case the mean value of maximum variation of L(t) is 0.3 nm while one for chemically non-connected cisplatin is 0.9 nm. At that a chemically connection of the heavy groups to the dendrimer ends cause some decrease of intramolecular mobility dendrimer-Pt. R, nm R, nm 3 ,5 2 ,0 3 ,0 1 ,5 2 ,5 2 ,0 1 ,0 1 ,5 1 ,0 0 ,5 0 100 200 300 400 500 0 T im e , p s 100 200 300 T im e , p s 400 500 a b Fig. 6. Time dependence of the distances L(t) between adsorbed (a) and chemically attached (b) cisplatins at 293 K. Acknowledgment The work was supported by ESF program SUPERNET, NWO (project 99 005 725), and INTAS (project 00-0712). 4 - 30 REFERENCES 1. Pil P., Lippard S. J. In Encyclopedia of Cancer, Ed. J. R. Bertino, Academic Press: San Diego, CA, 1997, Vol. 1, pp. 392-410. 2. Cohen S.M., Lippard S.J.: “Cisplatin: Fron DNA Damage to Cancer Chemotherapy.” Progr. Nucl. Acids Res. Mol. Biol. 2001 67 93-130. 3. Malik N., Evagorou E.G., Duncan R.: “Dendrimer-platinate: a novel approach to cancer chemotherapy.“ Anti-cancer Drugs 1999 10 (8) 767-75. 4. Gianasi E., Wasil M., Evagorou E.G., Keddle A., Wilson G., Duncan R.: “HPMA copolymer platinates as novel antitumour agents: in vitro properties, pharmacokinetics and antitumour activity in vivo.” Eur J Cancer 1999 35 (6) 9941002. 5. Esfand R., Tomalia D.A.: “Poly(amidoamine) (PAMAM) dendrimers: from biomimicry to drug delivery and biomedical applications.” Drug Discovery Today 2001 6 (8) 427-36. 6. Cloninger M.J.: “Biological applications of dendrimers.” Curr Opin Chem Biol. 2002 6 (6) 742-8. 7. Kolhe P., Misra E., Kannan R.M., Kannan S., Lieh-Lai M.: "Drug Complexation, in Vitro Release and Cellular Entry of Dendrimers and Hyperbranched Polymers." International Journal of Pharmaceutics 2003 259 (1-2) 143-60. 8. Beezer A.E., King A.S.H., Martin I.K., Mitchel J.C., Twyman L.J., Wain C.F.: "Dendrimers as Potential Drug Carriers; Encapsulation of Acidic Hydrophobes Within Water Soluble Pamam Derivatives." Tetrahedron 2003 59 (22) 3873-80. 9. Ghosh S.K, Kawaguchi S., Jinbo Y., Izumi Y, Yamaguchi K., Taniguchi T, Nagai K., Koyama K.: "Nanoscale Solution Structure and Transfer Capacity of Amphiphilic Poly(Amidoamine) Dendrimers Having Water and Polar Guest Molecules Inside." Macromolecules 2003 36 (24) 9162-9. 10. Lescanec R.L., Muthukumar M.: “Configurational characteristics and scaling behavior of starburst molecules: a computational study.” Macromol. 1990 23 (8) 2280-8. 11. Mansfield M.L., Klushin L.I.: “Intrinsic-Viscosity of Model Starburst Dendrimers.” J. Phys. Chem. 1992 96 (10) 3994-8. 12. Murat M., Grest G.S.: “Molecular-Dynamics Study of Dendrimer Molecules in Solvents of Varying Quality.” Macromolec. 1996 29 (4) 1278-85. 13. Karatasos K., Adolf D.B., Davies G.R.: “Statics and dynamics of model dendrimers as studied by molecular dynamics simulations.” J. Chem. Phys. 2001 115 (11) 5310-8. 14. Tomalia D.A., Naylor A.M., Goddard W.A., Kiefer G.E.: “Sturbust Dendrimers. Molecular Shape Control.” J. Am. Chem. Soc. 1989 111 (6) 2339-2341. 15. Miklis P., Cagin T.: “Goddard III W.A. Dynamics of Bengal Rose Encapsulated in the Meijer Dendrimer Box.” J. Am. Chem. Soc. 1997 119 (32) 7458-62. 16. Jahromi S., Coussens B., Meijerink N., Braam A.W.M.: “Side-Chain Dendritic Polymers - Synthesis and Physical-Properties.” J. Am. Chem. Soc. 1998 120 (38) 9753-62. 17. Mazo M.A., Zhilin P.A., Gusarova E.B., Sheiko S.S., Balabaev N.K.: “Computer simulation of intramolecular mobility of dendrimers.” J. Molec. Liqu. 1999 82 (23) 105-116. 18. Elshakre1 M., Atallah A.S., Santos S., Grigoras S.: “A structural study of carbosilane dendrimers versus polyamidoamine.” Computational and Theoretical Polymer Science 2000 10 (1) 21–28. 19. Gorman C.B., Smith J.C.: "Structure-Property Relationships in Dendritic Encapsulation." Accounts of Chemical Research 2001 34 (1) 60-71. 4 - 31 20. Lee I., Athey B.D., Wetzel A.W., Meixner W., Baker J.R.: "Structural Molecular Dynamics Studies on Polyamidoamine Dendrimers for a Therapeutic Application: Effects of Ph and Generation." Macromolecules 2002 35 (11) 4510-20. 21. Wilson M.R., Ilnytskyi J.M., Stimson L.M.: “Computer simulations of a liquid crystalline dendrimer in liquid crystalline solvents.” J. Chem. Phys. 2003 119 (6) 3509-15. 22. Paulo P.M.R., Costa S.M.B.: "Non-Covalent Dendrimer-Porphyrin Interactions: the Intermediacy of H-Aggregates?" Photochemical & Photobiological Sciences 2003 2 (5) 597-604. 23. Pricl S., Fermeglia M., Ferrone M., Asquini A.: "Scaling Properties in the Molecular Structure of Three- Dimensional, Nanosized Phenylene-Based Dendrimers as Studied by Atomistic Molecular Dynamics Simulations." Carbon 2003 41 (12) 2269-83. 24. Mazo M.A., Shamaev M.Yu., Balabaev N.K., Darinskii A.A., Neelov I.M.: “Conformational mobility of carbosilane dendrimer: Molecular Dynamics Simulation.” Phys. Chem. Chem. Phys. 2004 6 (6) 1285-9. 25. Canetta E., Maino G.: "Molecular Dynamic Analysis of the Structure of Dendrimers." Nuclear Instruments & Methods in Physics Research Section B- Beam Interactions With Materials and Atoms 2004 213 (1) 71-4. 26. Weiner S.J., Kollman P.A., Case D.A., Singh U.C., Ghio C., Alagona G., Profeta S., Weiner P.: “A new force field for molecular mechanical simulation of nucleic acids and proteins.” J. Am. Chem. Soc. 1984 106 (3) 765-84. 27. Jorgensen W.L., Chandrasekar J., Madura J.D., Impey R.W., Klein M.L.: “Comparison of simple potential functions for simulating liquid water.” J. Chem. Phys. 1983 79 (2), 926-35. 28. Cornell W.D., Cieplak P., Bayly C.L., Gould I.R., Merz Jr K.M.., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A.: “A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules.” J. Am. Chem. Soc. 1995 117 (19) 5179-97. 29. Yao S., Plastaras J.P., Marzilli L.G.: “A Molecular Mechanics AMBER-Type Force Field for Modeling Platinum Complexes of Guanine Derivatives.” Inorg. Chem. 1994 33 (26) 6061-77. 30. Allen M.P., Tildesley D.J.: Computer Simulation of Liquids. Oxford, Clarendon, 1987. 31. Lemak A.S., Balabaev N.K.: “On the Berendsen Thermostat.” Mol. Simul. 1995 15 223-31. 32. Berendsen H.J.C., Postma J.P.M., DiNola A., Haak J.R.: “Molecular dynamics with coupling to an external bath.” J. Chem. Phys. 1984 81 (8) 3684–90. 33. Balabaev N.K., Bessonov V.V., Neelov I.M., Mazo M.A.: “Computer simulation of dendrimers and their complexes with small guest molecules in water environment.” Abstr. Euroconf. SUPERNET 2004: Multiscale Phenomena in Material Structure Formation. Slovenia, 2004, p.13. 4 - 32
![Training Set Documents [1-100] 1. Agrawal A, Min DH, Singh N, Zhu](http://s3.studylib.net/store/data/006849311_1-841f76113ce605f46b23b81f034501c7-300x300.png)

![Test Set Documents [1-100] 1. Abderrezak A, Bourassa P](http://s3.studylib.net/store/data/006932013_1-fb7ae485eb54db57cb35fe73b994558f-300x300.png)
