Supporting Information - Royal Society of Chemistry
advertisement

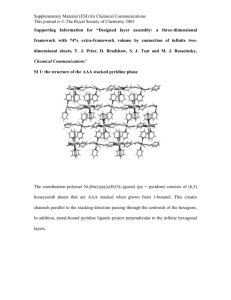
Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 SUPPORTING INFORMATION: Materials: Nucleic Acids. DNA polymers were purchased from Pharmacia Biotech, Inc. (Piscataway, NJ) and were used without further purification. DNA used include poly(dA), lot number: 9017836021; poly(dT), lot numbers: 8107834021 and 9107834021; poly(dA)(dT): lot number: 9067860021; Poly(rA): 7104110011, Poly(rU): lot number: 9034440021;. lot number: The concentrations of all the polymer solutions were determined spectrophotometrically using the following extinction coefficients (in units of mol of nucleotide/L)-1 cm-1= 8900 for poly(dA), = 9000 for poly(dT), = 6000 for poly(dA)•poly(dT), ), = 9800 for poly(rA), = 9350 for poly(rU).. Chemicals: Neomycin B, 1-pyrenebutyric acid N-hydroxysuccinimide ester, 2,4,6Triisopropylbenzenesulfonyl chloride, Di-tert-butyl dicarbonate, 2-aminoethanethiol hydrochloride, 4-dimethyl-aminopyridine and 1-aminopyrene were purchased from Acros (New Jersey) and were used without further purification. 4M HCl/disoxane was purchased from Aldrich and was used without further purification. Methylene chloride, DMF, dioxane were used as received. Pyridine was purified by CaH2 with distillation before using. Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 NHR O NH2 HO HO HO HO O H2 N NH2 O HO O O OH O HO HO NHR HO (Boc)2 O, DMF/H 2 O NH2 Et 3N, 5h, 60% HO HO NH2 O OH H2 N O RHN O O O NHR OH NHR O OH RHN 1a Neomycin B (1) TIBSCl, pyridine, rt, 40h, 60% NHR O HO HO NHR S NH2 HO HO RHN O HO HO NHR TIBSO NH2CH 2CH 2SH RHN O O O NHR O NHR OH NaOEt/EtOH, 12h, rt. 50% O HO HO NHR O OH RHN O O O NHR OH NHR O OH RHN 1c 1b O n NHR O O O O H N n n=3 O HO HO RHN S O HO HO CH 2Cl 2, rt, overnight, NHR OH NHR O OH RHN 80% RHN O O O n=3 1d 4M HCl/dioxane HSCH 2CH 2SH, 5min., rt, 80% Abbreviations R = Boc = di-tert-butyldicarbonate TPSCl = 2,4,6-triisopropylbenzenesulfonyl chloride NH2 H N n O HO HO O HO HO H2 N S NH2 O H2 N O O O OH NH2 O OH 1e Figure S1. Synthesis of Pyrene-Neomycin n=3 NH2 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 Pyrene-neomycin Synthesis -Experimental Compound 1a-1c have previously been reported (Kirk, S. R., Luedtke, N. W., Tor, Y., J. Am. Chem. Soc., 2000, 122, 980-981). Compound 1d. Compound 1c (70mg, 0.055mmol) and 4-dimethyl-aminopyridine (1mg) were dissolved in 5ml anhydrous methylene chloride at room temperature. 1pyrenebutyric acid N-hydroxysuccinimide ester (64mg, 0.165mmol, 3 equiv.) was added into the solution. The reaction mixture was kept stirring overnight. Organic solvent was then removed by vacuum. Flash column chromatography (5% CH3OH in CH2Cl2) afforded the desired product as a white solid. (62mg, 73%). Rf 0.48 (10% CH3OH in CH2Cl2); 1HNMR (500MHz, methanol-d4, 25C): 7.9-8.3 (m, 9H, Acr), 5.48 (br, 1H), 5.37 (1H), 5.13 (1H), 4.92 (1H), 4.23 (1H), 4.09(2H), 3.89 (2H), 3.76 (2H), 3.44-3.56 (6H), 3.0-3.3 (9H), 2.86 (2H), 2.7 (4H), 2.38 (2H), 2.18 (2H), 1.94 (1H), 1.4 (m, 54H). Compound 1e. Compound 1d (10mg) was dissolved in dioxane (0.6ml, treated over alumina). 1, 2-ethanedithiol (0.002 ml) was added followed by 4M HCl/dioxane (0.6ml). The solution was swirled for 5 min by hand upon which a pale yellow precipitate formed. Further precipitation was induced by adding ether and hexane (0.6ml each). Precipitate was recovered by centrifugation and several washes with ether and hexane. The paleyellow solid was redissolved in water and lyophilized overnight (6mg, 80% yield). max =342nm; 342 = 16778 cm-1M-1 C45H66N7O13S 944. HRMS (MALDI) m/z [M+2H]+ 946, calcd for Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 1 H NMR of 1a in CD3OD Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 1 H NMR of 1b in CDCl3 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 1 H NMR of 1c in CD3OD Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 1 H NMR of 1d in CD3OD Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 NH2 H N O HO HO H2 N 9.0 8.5 8.0 7.5 7.0 6.5 6.0 5.5 1 5.0 4.5 4.0 3.5 H NMR of 1e in D2O 3.0 2.5 2.0 O HO HO S NH2 O H2 N O O O NH2 OH NH2 O OH 1.5 1.0 0.5 0.0 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 MS (MALDI) Spectrum of 1e Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 2. 5 2.5 2 2 1. 5 A 1.5 1 1 0. 5 0.5 0 0 20 0 25 0 30 0 35 0 40 0 45 0 50 0 55 0 W av elength (nm) UV spectrum of Pyrene-neomycin 264 = 10956 cm-1M-1; 275 = 19152 cm-1M-1; 326 = 10663 cm-1M-1; 342 = 16778 cm-1M-1. Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 Melting Curves UV melting profiles of the poly(dA)2poly(dT) in the presence of 150mM KCl at the indicated ligand concentrations. [DNA]=15M base triplet. Solution conditions: 10 mM sodium cacodylate buffer, 0.5 mM EDTA, pH 7.2. Samples were heated from 20 to 90C at 5 deg/min, the annealing(90-5C) and the melting (5-90C) were conducted at 0.2 deg/min, and the samples were brought back to 20C at a rate of 5 deg/min. 0 M ligand 0.5 M 1-Aminopyrene 0.21 0.23 0.225 0.205 0.22 71 A 26 0 A 26 71 0 0.2 0.195 0.215 0.21 32 0.19 32 0.205 0.185 0.2 0.18 0.195 0 20 40 60 80 T( C) 100 20 1 M 1-Aminopyrene 30 40 50 60 T( C) 70 90 100 90 100 2 M 1-Aminopyrene 0.215 0.215 0.21 0.21 71.1 71.1 0.205 A 26 0 0 0.205 A 26 80 0.2 0.195 0.2 0.195 32 0.19 32.7 0.19 0.185 0.185 20 30 40 50 60 T( C) 70 80 90 100 20 30 40 50 60 T( C) 70 80 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 4 M 1-Aminopyrene 10 M 1-Aminopyrene 0.205 0.3 0.2 0.29 71.4 71 A 26 0.19 0.27 32.4 32.2 0.185 0.26 0.18 0.25 20 30 40 50 60 70 80 90 100 90 100 T( C) 20 M 1-Aminopyrene 0.31 0.3 0.29 71 0 A 26 0.28 A 26 0 0 0.195 0.28 0.27 32.4 0.26 0.25 20 30 40 50 60 T( C) 70 80 0 20 40 60 T( C) 80 100 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 UV melting profiles of the poly(dA)2poly(dT) in the presence of 150mM KCl at the indicated ligand concentrations. [DNA]=15M base triplet. Solution conditions: 10 mM sodium cacodylate buffer, 0.5 mM EDTA, pH 7.2. Samples were heated from 20 to 90C at 5 deg/min, the annealing(90-5C) and the melting (5-90C) were conducted at 0.2 deg/min, and the samples were brought back to 20C at a rate of 5 deg/min. polydA2polydT with 0.5 M pyrene-7-neo polydA2polydT with 0 M ligand 0.3 0.32 0.28 0.3 A260 A260 70 0.26 0.24 0.26 36.5 32 0.22 71 0.28 0.24 0.2 0.22 20 30 40 50 o 60 T( C) 70 80 90 20 30 polydA2polydT with 1M pyrene-7-neo 0.32 0.3 0.3 71 A260 A260 60 T( o C) 70 80 90 72.2 0.28 0.26 43.3 0.24 50 polydA2polydT with 2M pyrene-7-neo 0.32 0.28 40 0.26 55.6 0.24 0.22 0.22 0.2 0.2 20 30 40 50 o 60 T( C) 70 80 90 20 polydA2poldT with 4M pyrene-7-neo 30 40 50 60 T( o C) 70 80 90 polydA2polydT with 6M pyrene-7-neo 0.47 0.34 0.46 0.32 0.45 72.6 A260 A260 0.3 0.28 59.1 0.26 74.2 0.44 0.43 0.42 62 0.41 0.24 0.4 0.39 0.22 20 30 40 50 o 60 T( C) 70 80 90 30 40 50 60 T( o C) 70 80 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 UV melting profiles of the poly(dA)2poly(dT) in the presence of 150mM KCl at the indicated ligand concentrations. [DNA]=15M base triplet. Solution conditions: 10 mM sodium cacodylate buffer, 0.5 mM EDTA, pH 7.2. Samples were heated from 20 to 90C at 5 deg/min, the annealing(90-5C) and the melting (5-90C) were conducted at 0.2 deg/min, and the samples were brought back to 20C at a rate of 5 deg/min. polydA2polydT with 0.5 M neomycin polydA2polydT with 0 M ligand 0.42 0.44 0.4 0.42 71 0.4 A260 A260 0.38 0.36 71 0.38 0.36 0.34 33 34 0.34 0.32 0.32 0.3 24 32 40 48 56 T ( o C) 64 72 24 80 0.42 0.42 0.4 0.4 71 A260 A260 48 56 T ( o C) 0.38 0.36 0.34 40 64 72 80 polydA2polydT with 2M neomycin polydA2polydT with 1M neomycin 0.38 32 71 0.36 0.34 35.1 36.5 0.32 0.32 0.3 0.3 24 32 40 48 56 T ( o C) 64 72 80 24 polydA2polydT with 4M neomycin 32 40 48 56 T ( o C) 64 72 80 72 80 polydA2polydT with 6M neomycin 0.36 0.5 0.48 0.34 0.46 72.1 0.44 A260 A 260 0.32 0.3 72.2 0.42 0.4 0.28 0.38 43 0.26 46 0.36 0.24 0.34 24 32 40 48 o 56 T( C) 64 72 80 24 32 40 48 56 T ( o C) 64 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 UV melting profiles of the poly(dA)2poly(dT) in the presence of 150mM KCl at the indicated ligand concentrations. [DNA]=15M base triplet. Solution conditions: 10 mM sodium cacodylate buffer, 0.5 mM EDTA, pH 7.2. Samples were heated from 20 to 90C at 5 deg/min, the annealing(90-5C) and the melting (5-90C, curves shown below) were conducted at 0.2 deg/min, and the samples were brought back to 20C at a rate of 5 deg/min. polydA2polydT with 0.5 M neo + 0.5 M1-aminopyrene polydA2polydT with 0 M ligand 0.42 0.42 0.4 0.4 71 0.38 A260 A260 0.38 0.36 0.34 71 0.36 0.34 33 34.5 0.32 0.32 0.3 0.3 24 32 40 48 o 56 64 72 80 24 32 40 48 56 64 72 80 T ( C) T ( o C) polydA2polydT with 1 M neo + 1 M 1-aminopyrene polydA2polydT with 2 M neo + 2 M 1-aminopyrene 0.42 0.5 0.48 0.4 0.46 71 72.8 0.44 A260 A260 0.38 0.36 0.42 0.4 0.34 36 0.38 40 0.32 0.36 0.3 0.34 24 32 40 48 56 T ( o C) 64 72 80 24 32 40 48 56 T ( o C) 64 72 80 polydA2polydT with 6 M neo + 6 M 1-aminopyrene polydA2polydT with 4 M neo +4 M 1-aminopyrene 0.42 0.42 0.4 0.4 71 71 A260 0.38 A260 0.38 0.36 53 0.36 49 43 49 0.34 0.34 0.32 0.32 24 32 40 48 56 T ( o C) 64 72 80 24 32 40 48 56 T ( o C) 64 72 80 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 UV melting profiles (287nm) of the poly(rA)poly(rU) and poly(rA)2poly(rU) in the presence of 35mM NaCl at the indicated ligand concentrations. [RNA]=30M base triplet. Solution conditions: 10 mM sodium cacodylate buffer, 0.1 mM EDTA, pH 6.8. Samples were heated from 20 to 90C at 0.2 deg/min. polyrApolyrU with 0 M ligand polyrA2polyrU with 0 M ligand 0. 32 0. 35 0. 315 0. 31 A 287 48 A 287 48.0 0. 305 0. 3 0. 295 0. 29 0. 285 0. 28 0. 3 20 25 30 35 40 45 o 50 55 60 20 25 30 35 40 o 45 50 55 60 T( C) T( C) polyrApolyrU with 1 M pyrene-7-neo polyrA2polyrU with 1 M pyrene-7-neo 0. 33 0. 35 0. 325 0. 34 0. 32 A 287 A 287 48 0. 315 0. 31 0. 305 48 0. 33 0. 32 0. 3 0. 31 0. 295 0. 29 0. 3 20 25 30 35 40 o 45 50 55 60 20 25 30 35 T( C) 40 45 T( oC) 50 55 60 polyrA2polyrU with 2 M pyrene-7-neo polyrApolyrU with 2 M pyrene-7-neo 0. 33 0. 35 0. 345 0. 32 49.2 0. 31 A 287 A 287 0. 34 50 0. 335 0. 3 0. 33 0. 325 0. 32 0. 29 0. 315 0. 28 0. 31 20 25 30 35 40 45 T( oC) 50 55 60 20 25 30 35 40 45 T( oC) 50 55 60 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 polyrA2polyrU with 4 M pyrene-7-neo polyrApolyrU with 4 M pyrene-7-neo 0. 35 0. 37 0. 34 0. 36 54.7 0. 35 A 287 A 287 0. 33 55.2 0. 32 0. 34 0. 31 0. 33 0. 3 0. 32 0. 29 0. 31 20 30 40 o50 T( C) 60 70 20 30 40 o 50 T( C) 60 70 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 UV melting profiles (280nm) of the poly(rA)poly(rU) and poly(rA)2poly(rU) in the presence of 35mM NaCl at the indicated ligand concentrations. [RNA]=30M base triplet. Solution conditions: 10 mM sodium cacodylate buffer, 0.1 mM EDTA, pH 6.8. Samples were heated from 20 to 90C at 0.2 deg/min. polyrApolyrU with 0 M ligand polyrA2polyrU with 0 M ligand 0. 354 0. 45 0. 352 0. 445 31 A 260 A 280 0. 35 0. 348 0. 346 0. 44 0. 435 0. 344 0. 43 0. 342 0. 34 0. 425 20 25 30 35 o 40 45 50 20 25 30 35 40 45 50 T( C) T( oC) polyrApolyrU with 1 M pyrene-7-neo polyrA2polyrU with 1 M pyrene-7-neo 0. 364 0. 445 0. 362 0. 44 A 280 A 280 0. 36 0. 358 34.1 0. 435 0. 356 0. 43 0. 354 0. 352 0. 425 20 25 30 35 o 40 45 50 20 25 30 35 o 40 45 50 T( C) T( C) polyrApolyrU with 2 M pyrene-7-neo polyrA2polyrU with 2 M pyrene-7-neo 0. 36 0. 46 0. 455 0. 358 A 280 A 280 0. 45 0. 356 0. 354 0. 445 36.1 0. 44 0. 435 0. 352 0. 43 0. 35 0. 425 20 25 30 35 o T( C) 40 45 50 20 25 30 35 o T( C) 40 45 50 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 polyrA2polyrU with 4 M pyrene-7-neo polyrApolyrU with 4 M pyrene-7-neo 0. 39 0. 49 0. 48 A 280 A 280 0. 385 0. 38 0. 47 40.1 0. 46 0. 375 0. 45 0. 37 0. 44 20 25 30 35 40 o T( C) 45 50 55 60 20 25 30 35 40 o T( C) 45 50 55 60 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 UV melting profiles (284nm) of the poly(rA)poly(rU) and poly(rA)2poly(rU) in the presence of 35mM NaCl at the indicated ligand concentrations. [RNA]=30M base triplet. Solution conditions: 10 mM sodium cacodylate buffer, 0.1 mM EDTA, pH 6.8. Samples were heated from 20 to 90C at 0.2 deg/min. polyrA2polyrU with 0 M ligand 0. 35 0. 4 0. 34 0. 39 0. 33 0. 38 A 284 A 284 polyrApolyrU with 0 M ligand 48 0. 32 48 0. 37 0. 31 0. 36 0. 3 0. 35 0. 29 31.7 0. 34 20 25 30 35 o 40 T( C) 45 50 55 60 20 polyrApolyrU with 1 M pyrene-7-neo 25 30 35 40 T( oC) 45 50 55 60 polyrA2polyrU with 1 M pyrene-7-neo 0. 35 0. 39 0. 385 34.9 0. 38 48 48.0 A 284 A 284 0. 375 0. 37 0. 365 0. 36 0. 355 0. 3 0. 35 20 25 30 35 40 45 o 50 55 60 20 25 30 35 40 45 o 50 55 60 T( C) T( C) polyrApolyrU with 2 M pyrene-7-neo polyrA2polyrU with 2 M pyrene-7-neo 0. 35 0. 395 0. 39 36.1 0. 385 A 284 A 284 0. 38 50 50.4 0. 375 0. 37 0. 365 0. 36 0. 3 0. 355 20 30 40 o 50 T( C) 60 70 20 30 40 o 50 T( C) 60 70 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 polyrA2polyrU with 4 M pyrene-7-neo polyrApolyrU with 4 M pyrene-7-neo 0. 37 0. 42 0. 36 0. 41 40.1 55 55.2 0. 4 A 284 A 284 0. 35 0. 34 0. 39 0. 33 0. 38 0. 32 0. 37 0. 31 0. 36 20 30 40 o 50 T( C) 60 70 20 30 40 o 50 T( C) 60 70 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 UV melting profiles (280nm) of the poly(rA)poly(rU) and poly(rA)2poly(rU) in the presence of 35mM NaCl at the indicated ligand concentrations. [RNA]=30M base triplet. Solution conditions: 10 mM sodium cacodylate buffer, 0.1 mM EDTA, pH 6.8. Samples were heated from 20 to 90C at 0.2 deg/min. polyrA2polyrU with 0 M ligand 0. 36 0. 45 0. 355 0. 445 A 280 A 280 polyrApolyrU with 0 M ligand 0. 35 0. 345 35 0. 44 0. 435 0. 34 0. 43 20 25 30 35 40 o 45 50 20 25 30 35 40 o T( C) 45 50 T( C) polyrApolyrU with 1 M neomycin polyrA2polyrU with 1 M neomycin 0. 358 0. 445 0. 356 0. 44 A 280 A 280 0. 354 0. 352 0. 35 0. 348 41 0. 435 0. 43 0. 346 0. 344 0. 425 20 24 28 32 36 40 o 44 48 52 20 24 28 32 T( C) polyrApolyrU with 2 M neomycin 40 o 44 48 52 polyrA2polyrU with 2 M neomycin 0. 375 0. 46 0. 37 0. 45 0. 365 0. 44 A 280 A 280 36 T( C) 0. 36 0. 355 43.6 0. 43 0. 42 0. 35 0. 41 20 25 30 35 o 40 T( C) 45 50 55 20 25 30 35 o 40 T( C) 45 50 55 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 polyrA2polyrU with 4 M neomycin 0. 344 0. 44 0. 342 0. 435 0. 34 0. 43 A 280 A 280 polyrApolyrU with 4 M neomycin 0. 338 0. 336 0. 334 0. 425 0. 42 46 0. 415 0. 332 0. 41 0. 33 0. 405 20 25 30 35 40 o T( C) 45 50 55 60 30 35 40 45 o T( C) 50 55 60 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 UV melting profiles (284nm) of the poly(rA)poly(rU) and poly(rA)2poly(rU) in the presence of 35mM NaCl at the indicated ligand concentrations. [RNA]=30M base triplet. Solution conditions: 10 mM sodium cacodylate buffer, 0.1 mM EDTA, pH 6.8. Samples were heated from 20 to 90C at 0.2 deg/min. polyrA2polyrU with 0 M ligand 0.4 0. 33 0.39 48 0. 32 A284 A 284 polyrApolyrU with 0 M ligand 0. 34 0. 31 34.5 0.38 48.8 0.37 0.36 0. 3 0.35 0. 29 20 25 30 35 40 o 45 50 55 20 60 25 30 35 T( C) 40 45 T( o C) 50 55 60 polyrA2polyrU with 1 M neomycin polyrApolyrU with 1 M neomycin 0.395 0. 35 41 0.39 0. 34 51.2 0.38 51.2 0. 33 A284 A 284 0.385 0. 32 0.375 0.37 0.365 0. 31 0.36 0.355 0. 3 20 25 30 35 40 45 o 50 55 20 60 25 30 35 T( C) polyrApolyrU with 2 M neomycin 40 45 T( o C) 50 55 60 polyrA2polyrU with 2 M neomycin 0.36 0.39 42.6 0.385 0.35 A284 A284 0.38 52 0.34 0.33 51.6 0.375 0.37 0.365 0.32 0.36 0.31 0.355 20 25 30 35 40 o 45 T( C) 50 55 60 20 25 30 35 40 45 T( o C) 50 55 60 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 polyrApolyrU with 4 M neomycin polyrA2polyrU with 4 M neomycin 0.335 0.4 0.33 53.2 0.325 0.39 A284 A284 0.32 0.315 0.31 0.38 53.2 0.37 0.305 0.36 0.3 0.295 0.35 20 25 30 35 40 o 45 T( C) 50 55 60 28 32 36 40 44 48 T( o C) 52 56 60 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 UV melting profiles (287nm) of the poly(rA)poly(rU) and poly(rA)2poly(rU) in the presence of 35mM NaCl at the indicated ligand concentrations. [RNA]=30M base triplet. Solution conditions: 10 mM sodium cacodylate buffer, 0.1 mM EDTA, pH 6.8. Samples were heated from 20 to 90C at 0.2 deg/min. polyrA2polyrU with 0 M ligand polyrApolyrU with 0 M ligand 0.32 0.35 0.315 0.34 0.31 A287 A287 0.305 48 0.3 0.295 48.8 0.33 0.32 0.29 0.31 0.285 0.28 0.3 20 25 30 35 40 o 45 50 55 60 20 25 30 35 T( C) 0.325 0.345 0.32 0.34 0.315 0.335 A287 A287 0.35 51.2 0.31 45 50 55 60 polyrA2polyrU with 1 M neomycin polyrApolyrU with 1 M neomycin 0.33 0.305 40 T( o C) 0.33 0.325 0.3 0.32 0.295 0.315 0.29 51.2 0.31 20 25 30 35 40 o 45 T( C) 50 55 60 20 polyrApolyrU with 2 M neomycin 25 30 35 40 45 T( o C) 50 55 60 polyrA2polyrU with 2 M neomycin 0.34 0.35 0.345 52 0.34 51.2 0.335 0.32 A287 A287 0.33 0.31 0.33 0.325 0.32 0.3 0.315 0.29 0.31 20 25 30 35 40 o 45 T( C) 50 55 60 20 25 30 35 40 45 T( o C) 50 55 60 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 polyrApolyrU with 4 M neomycin polyrA2polyrU with 4 M neomycin 0.32 0.37 0.315 0.36 0.31 0.35 53 0.3 A287 A287 0.305 0.295 53.1 0.34 0.33 0.29 0.32 0.285 0.28 0.31 20 30 40 o 50 T( C) 60 70 30 35 40 45 50 55 T( o C) 60 65 70 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 UV melting profiles (280nm) of the poly(rA), poly(rU) and poly(rA)2poly(rU) in the presence of 35mM NaCl at the indicated ligand concentrations. [RNA]=30M base triplet. Solution conditions: 10 mM sodium cacodylate buffer, 0.1 mM EDTA, pH 6.8. Samples were heated from 20 to 90C at 0.2 deg/min. polyrA2polyrU with 0 M ligand polyrApolyrU with 0 M ligand 0.33 0.435 0.329 48 0.328 0.43 A280 A280 0.327 0.326 0.325 35 0.425 0.42 0.324 0.415 0.323 0.322 0.41 20 25 30 35 40 o 45 T( C) 50 55 60 20 25 polyrApolyrU with 1M 1-aminopyrene 30 35 40 45 T( o C) 50 55 60 55 60 55 60 polyrA2polyrU with 1M 1-aminopyrene 0.35 0.445 0.348 0.44 48 0.346 A280 A280 0.344 0.342 0.34 35.7 0.435 0.43 0.338 0.425 0.336 0.334 0.42 20 25 30 35 40 o 45 T( C) 50 55 60 20 25 polyrApolyrU with 2M 1-aminopyrene 30 35 40 45 T( o C) 50 polyrA2polyrU with 2M 1-aminopyrene 0.355 0.445 0.44 49 0.435 A280 A280 0.35 0.345 35.7 0.43 0.425 0.34 0.42 0.335 0.415 20 25 30 35 40 o 45 T( C) 50 55 60 20 25 30 35 40 45 T( o C) 50 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 polyrApolyrU with 4M 1-aminopyrene 0.35 0.48 0.345 0.47 35.4 48 A280 0.34 A280 polyrA2polyrU with 4M 1-aminopyrene 0.335 0.46 0.45 0.33 0.44 0.325 0.32 0.43 20 25 30 35 40 o 45 T( C) 50 55 60 20 25 30 35 40 45 T( o C) 50 55 60 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 UV melting profiles (284 nm) of the poly(rA), poly(rU) and poly(rA)2poly(rU) in the presence of 35mM NaCl at the indicated ligand concentrations. [RNA]=30M base triplet. Solution conditions: 10 mM sodium cacodylate buffer, 0.1 mM EDTA, pH 6.8. Samples were heated from 20 to 90C at 0.2 deg/. polyrA2polyrU with 0 M ligand 0.38 0.31 0.37 48 0.3 A284 A284 polyrApolyrU with 0 M ligand 0.32 0.29 0.28 34 48 0.36 0.35 0.34 0.27 0.33 20 25 30 35 40 o 45 T( C) 50 55 60 20 25 polyrApolyrU with 1M 1-aminopyrene 30 35 40 45 T( o C) 50 60 polyrA2polyrU with 1M 1-aminopyrene 0.34 0.39 0.33 0.38 34.5 48 0.37 0.32 A284 A284 55 0.31 0.36 0.35 0.3 0.34 0.29 0.33 20 25 30 35 40 45 T( o C) 50 55 60 20 25 polyrApolyrU with 2M 1-aminopyrene 30 35 40 45 T( o C) 50 55 60 55 60 polyrA2polyrU with 2M 1-aminopyrene 0.34 0.39 0.33 0.38 0.32 0.37 A284 A284 35 0.31 0.3 48 0.36 0.35 0.29 0.34 20 25 30 35 40 o 45 T( C) 50 55 60 20 25 30 35 40 45 T( o C) 50 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 polyrA2polyrU with 4M 1-aminopyrene polyrApolyrU with 4M 1-aminopyrene 0.34 0.43 35 0.42 0.33 0.41 48 0.4 A284 A284 0.32 48.1 0.31 0.3 0.39 0.38 0.37 0.29 0.36 0.28 0.35 20 25 30 35 40 o 45 T( C) 50 55 60 20 25 30 35 40 45 T( o C) 50 55 60 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 UV melting profiles (287 nm)of the poly(rA), poly(rU) and poly(rA)2poly(rU) in the presence of 35mM NaCl at the indicated ligand concentrations. [RNA]=30M base triplet. Solution conditions: 10 mM sodium cacodylate buffer, 0.1 mM EDTA, pH 6.8. Samples were heated from 20 to 90C at 0.2 deg/min and the samples were brought back to 5C at a rate of 5 deg/min. polyrA2polyrU with 0 M ligand 0.33 0.295 0.325 0.29 0.32 0.285 0.315 0.28 A287 A287 polyrApolyrU with 0 M ligand 0.3 48 0.275 0.305 0.27 0.3 0.265 0.295 0.26 48.8 0.31 0.29 20 25 30 35 40 o 45 T( C) 50 55 60 20 25 35 40 45 T( o C) 50 55 60 55 60 55 60 polyrA2polyrU with 1M 1-aminopyrene 0.32 0.34 0.31 0.33 0.3 A287 A287 polyrApolyrU with 1M 1-aminopyrene 30 48.1 0.29 0.28 48.4 0.32 0.31 0.3 0.27 0.29 20 25 30 35 40 o 45 50 55 60 20 25 30 35 T( C) polyrApolyrU with 2M 1-aminopyrene 40 45 T( o C) 50 polyrA2polyrU with 2M 1-aminopyrene 0.315 0.34 0.31 0.33 0.305 48.1 A287 A287 0.3 0.295 0.29 0.32 49 0.31 0.285 0.3 0.28 0.275 0.29 20 25 30 35 40 o 45 T( C) 50 55 60 20 25 30 35 40 45 T( o C) 50 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 polyrApolyrU with 4M 1-aminopyrene polyrA2polyrU with 4M 1-aminopyrene 0.31 0.37 0.36 0.3 A287 A287 0.35 48 0.29 0.28 48.4 0.34 0.33 0.32 0.27 0.31 0.26 0.3 20 25 30 35 40 o 45 T( C) 50 55 60 20 25 30 35 40 45 T( o C) 50 55 60 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2001 Computer Modeling Step I: DNA (TAT) triplex conformation The original structure of dT10dA10dT10 triplex was obtained from the NMR structure reported by Feigon (Biochemistry 1998). 27 sodium ions were added to neutralize the structure. The conformation was then optimized by AMPER* force field, in water, to within a gradient of 0.5 kJ/molÅ. The movements of the atoms of the external bases were restricted during minimization. Step II: Neomycin conformation Neomycin structure was built and optimized with MacroModel and SYBYL programs using AMBER* force field. The all atom AMBER* force field has been used since it contains parameters for both neomycin and the DNA triplex. In accordance with NMR experiments, five of the six-neomycin amines were protonated. Conformational searching was done with Monte Carlo routine from MacroModel, using all atom AMBER* force field, the force field charges, and the continuum GB/SA model of water as implemented in MacroModel. All the flexible bonds were selected. Conformation of neomycin bound to W-H groove. Step III Pyrene-neomycin conformation Pyrene-neomycin conformation was generated based on neomycin conformation. The generated conformation was then optimized by AMPER* force field and water as a solvent. Step IV Docking After optimizing the pyrene-neomycin conformation, the structure was docked into Waston-Hoogsteen groove of TAT triplex in different orientations. Restricting the movement of the triplex, the structure of the complex was optimized using AMBER* force field and water as a solvent.