Computational Neuroscience

Biology 450 / IBS 534 Computational Neuroscience Spring 2014
Course Syllabus and Class Schedule
Time and Locations:
Biology 450 / IBS 534 is taught Tuesday and Thursday from 4:00 p.m. to 5:30 p.m. at the Emory
Campus. We meet in room 109 Dental School (1462 Clifton). Session generally consist of a 30-60 min lecture part followed by computer exercises.
Evening Sessions:
For the graduate section of the class, there will be mandatory evening Journal club sessions every 2nd
Tuesday after class with Pizza. Once in a while there will be an illustrious guest speaker, but for the most part this is a Journal club where the graduate students in the class earn part of their grade to a widely invited Computational Neuroscience Audience. Pizza will be free.
Instructors:
Dr. Dieter Jaeger (course director), Professor, Dept. Biology, Emory University
Room 2129 Rollins Res. phone 404 727 8139, e-mail djaeger@emory.edu
Dr. Ron Calabrese, Professor, Dept. Biology, Emory University
Room 2167 Rollins Res. phone 404 727 0319, e-mail ronald.calabrese@emory.edu
Dr. Astrid Prinz, Associate Professor, Dept. Biology, Emory University
Room 2105 Rollins Res. Phone 404 727 9381, e-mail astrid.prinz@emory.edu
Grading
There will be 13 weekly homework assignments, of which the best 11 grades will count for 75% of the class grade. 25% of the grade is determined by class participation. For the graduate students, this participation grade includes one oral journal club presentation. All materials turned in must be typed
(except equations, drawings). The submission will be electronic via Blackboard. Each homework will have a deadline one week after it is received, unless noted otherwise by the instructor. The grade of late homeworks will be discounted by 3%. Homeworks will no longer be accepted after they are returned for other students.
Text
The recommended text for Bio450/IBS534 is:
Computational Modeling Methods for Neuroscientists, edited by Erik De Schutter, MIT Press,
2009, approx.. $45 [via amazon.com sellers as of 11/13]
The following manual for some GENESIS based tutorials is freely available on-line:
The Book of GENESIS: Exploring Realistic Neural Models with the GEneral NEural SImulation
System. b y James M. Bower, D. Beeman, 2 nd
edition, Springer 1998, AVAILABLE ONLINE AT: www.genesis-sim.org
Biology 450 / IBS 534 Computational Neuroscience Spring 2014
The following book is highly recommended as reference material for interested students:
Foundations of cellular neurophysiology , Johnston and Wu, MIT press, 1995, $77.53 [amazon.com]
Other Useful Reading Material (purchase only recommended in special cases).
A) General Neuroscience
Principles of Neuroscience , 4 th
edition, Kandel, Schwartz, Jessell, McGraw Hill, 2000
B) Detailed Cellular Neurophysiology
Ionic channels of excitable membranes , 3 rd
edition, B. Hille, Sinauer, 2001
C) Computational Neuroscience
Methods in neuronal modeling – from ions to networks . 2 nd
edition, C. Koch and I. Segev, MIT press, 1998
Biophysics of Computation – information processing in single neurons . C. Koch, Oxford Press,
1999,
Spikes . Rieke, Warland, van Steveninck, and Bialek, MIT press, 1997, $25.00 (paperback)
The theoretical foundation of dendritic function – selected papers of W. Rall . Segev, Rinzel,
Shepherd, MIT press, 1995
Dendrites . Stuart, Spruston, Hausser, Oxford U. Press., 1999
Theoretical Neuroscience . P. Dayan and L. Abbott. MIT Press, $50.00
D) Neural Networks
Computational Explorations in Cognitive Neuroscience: Understanding the Mind by Simulating the Brain (w/ CD).
O’Reilly, Munakata, McClelland. MIT press 2000. 2nd edition is available freely on-line
Biology 450 / IBS 534 Computational Neuroscience Spring 2014
Tues 2-18 11
Thur 2-20
HW6B
12
Tues 2-25
BONUS
Tues 3-4
HW 7
13
13a
Thur 2-27 14
15
Thur 3-6 16
Tues 3-18 17
HW 8
17a
Thur 3-20 18
Tues 3-25
HW9
19
Thur 3-27 20
Tues 4-1 21
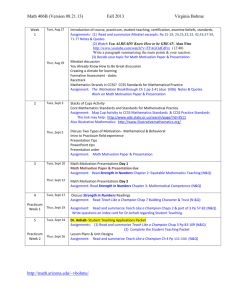
Class Schedule
Date
Tues 1-14
Thur 1-16
Tues1-21
Thur 1-23
Session Teacher
1
2
3
4
Tues 1-28 5
Thur 1-30
Tues 2-4
5a
6
7
Jae
Cal
Cal
Cal
Cal tba
Cal
Prinz
Lecture Description
Numerical Methods and Introduction to GENESIS
A passive compartment: R, C, and I Passive tutorial
Hodgkin Huxley 1 Squid tutorial
Hodgkin Huxley 2. Squid tutorial
Beyond Na and K: effects of conductances, e.g. bursting, plateau
Journal Club Session 1
Channel effects part II.
Computer exercise
Description
Passive tutorial
Single compartment tutorial with various channels
Single compartment tutorial with various channels xpp
Thur 2-6
Tues 2-11
8
9
Prinz
Cal
Dynamical Systems 1: Phase space and limit cycles
Dynamical Systems 1: Phase space and limit cycles
Calcium and molecular processes xpp
Thur 2-13
9a
10 tba
Jae
Journal Club Session 2
Molecular pathway simulations
Compartment w/ diffusion shell, Ca(K) – Burster
Tutorial kinetikit (genesis)
Jae
Jae
Cal
Cal tba
Jae
Jae
Jae tba
Cal
Cal
Prinz
Prinz
Single ion channel simulations Neuron single channel tut
Simulating Synapses: EPSPs and
IPSPs
Simulating the in vivo high conductance state
Journal Club Session 3
Cable theory: Lecture I and II
Cable theory: Lab
Modified Neuron tutorial
(GENESIS) none
Neuron and Neurokit
Small networks – pattern generation Leech heartbeat CPG
SPRING BREAK
Small networks – intersegmental Leech heartbeat CPG coordination
Journal Club Session 4
Realistic single neuron reconstructions Neurokit
Ion channels in spatially complex cells: Attenuation and dendritic spiking
Globus Pallidus Neuron
(GENESIS)
Introduction to large parameter spaces t.b.a.
Querying and data mining databases t.b.a.
Biology 450 / IBS 534 Computational Neuroscience Spring 2014
HW10
Thur 4-3 22
Tues 4-8
HW11
Tues 4-15
HW12
Tues 4-22
HW13
21a
23
Thur 4-10 24
25
25a
Thur 4-17 26
27
Thur 4-24 28
Tues 4-29 29a tba
Jae
Jae
Jae
Jae tba
Jae
Jae
Jae tba of large parameter searches
Journal Club Session 5
The Purkinje cell : Dendritic Plateau
Potential
The Purkinje cell: Synaptic
Integration
The Cerebellar nuclei: Control by inhibitory input
The Cerebellar nuclei: Behavioral activity Modulation
Journal Club Session 6
Simulating Short Term Synaptic
Plasticity
Simulating Long Term Synaptic
Plasticity
Olfactory Cortex and Oscillations
Journal Club Session 7
Purkinje tutorial
Purkinje tutorial
Genesis CN neuron simulation
Genesis CN neuron simulation
Genesis CN neuron simulation
Synfire. piriform tutorial