PSI+
advertisement

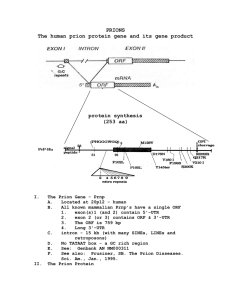
The yeast prion [PSI+]. Citation from Susan Lindquist, Member, Whitehead Institute Professor of Biology Investigator, Howard Hughes Medical Institute: Prions and Protein-based Inheritance: Prions are proteins that can acquire selfperpetuating changes in structure that alter protein function and cell phenotype. They represent an epigenetic mechanism of inheritance because the altered phenotype is passed from generation to generation through the heritable changes in protein structure, with no underlying changes in nucleic acids. We have shown by cell biological, genetic and biochemical data that this protein-only mechanism controls the inheritance of the yeast prion [PSI+]. [PSI+] is propagated through a self-perpetuating change in the conformational state of the translation termination factor Sup35p, which forms an amyloid structure in its [PSI+] prion state. Most recently we have been examining how many other proteins (in S. cerevisiae and C. elegans) can produce heritable switches in conformation and function, and whether this mechanism can serve as a new, general method for manipulating phenotypes. An extraordinary phenomenon of [PSI+] that we have discovered is its ability to mask a vast store of phenotypic variation. In more than 150 assays, conversion from [psi-] to [PSI+] provided the means to uncover a wide variety of hidden phenotypes and produce new, sometimes beneficial, heritable phenotypes in multiple genetic backgrounds. We suggest that the epigenetic and metastable nature of [PSI+] inheritance allows yeast cells to exploit pre-existing genetic variation to thrive in fluctuating environments without necessitating a change in genotype (see figure). Further, the capacity of [PSI+] to convert previously neutral genetic variation to a non-neutral state may facilitate the evolution of new traits. To further understand the mechanism of prion amyloid formation and transmission, we created a large number of cysteine-containing variants and employed fluorescent tags, crosslinking, and other methods to determine the nature of Sup35p's cooperatively folded amyloid core. We identified specific segments that form intermolecular contacts in a 'Head-to-Head', 'Tail-to-Tail' fashion, while a central region forms exclusively intramolecular contacts. This provided the first definition of the contacts that hold any amyloid fiber together. These structural insights allowed us to attack two central questions in prion biology, the mechanism of nucleation and the structural basis of prion strains. We established that the Head region acquires amyloidogenic interactions first and that these are sufficient to nucleate assembly. Prion strains, in both yeast and mammals, are distinct physical forms that produce different prion phenotypes in vivo. We found that variations in the length of the amyloid core and the nature of intermolecular interfaces were both characteristic of distinct strains of [PSI+] and sufficient to create them anew. Krishnan, R., Lindquist, S. L. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435:765-72. (2005). True, H., Berlin, I., Lindquist, S., Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 431:184-7. (2004). Information related to research project Prion are misshapen proteins that exist in at least two different conformations, one of which is self-replicating and expresses unique biological function are hypothesized to infect and propagate by refolding abnormally into a structure which is able to convert normal molecules of the protein into the abnormally structured form [PSI+] is the prion form of the translational release factor eRF3, encoded by the chromosomal gene SUP35, which suppresses nonsense codons Conformational change impairs the termination activity of eRF3 increasing stop codon readthrough, resulting in the production of proteins with carboxy terminus extensions [PSI+]-induced defect in translation termination could stimulate expression of frameshifted genes by affecting the reading frame maintenance of the ribosome, particularly if a codon particularly prone to allow +1 slippage is followed by a poorly recognized termination codon The only gene known to use a shifty-stop in yeast is the OAZ1