Mitosis, meiosis and Mendelian inheritance - Wk 1-2
advertisement

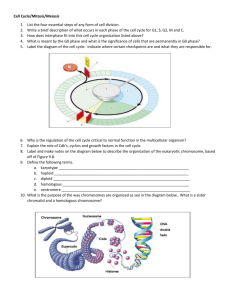
Mitosis, meiosis and Mendelian inheritance 1. Review meiosis and how it differs from mitosis Mitosis: Mitosis can be divided into five stages interphase, prophase, metaphase, anaphase and telophase. 1. Interphase: The total period from cell formation to cell division. G1 phase (Growth 1)- Cells are metabolically active, synthesize proteins rapidly and grow vigorously. As G1 ends the centrioles start to replicate in preparation for cell division. G0 phase (Growth 0)- cells that permanently ceased division are in this stage. S phase (Synthetic)- DNA replicates itself. New histones are made and assembled into chromatin. G2 phase (Growth 2)- Enzymes and proteins needed for division are synthesized and moved to their proper sites. By the end of G2 centriole replication is completed. 2. Late in interphase (G1) the chromosomes have already duplicated during S phase and appear as a dark mass in the nucleus and are not individually distinguishable. The two centrosomes also appear. 3. Prophase: Chromatin fibres become visible and the centrosomes migrate to opposite ends of the cell forming the mitotic spindle. Each duplicated chromosome becomes visible and appears as two identical sister chromatids joined at the centromere. 4. Metaphase: The nuclear envelope fragments allowing the microtubules of the mitotic spindle to interact with the chromosomes. The chromosomes align in the centre of the cell on the microtubules with centrosomes at opposite poles. 5. Anaphase: The paired centromeres divide releasing the sister chromosomes and the individual chromosomes migrate to opposite poles toward the centrosomes due to microtubules shortening. 6. Telophase and Cytokinesis: Nuclear envelopes begin to form around the chromosomes at each end of the cell from fragments of the parent cell’s nuclear envelope. In the process of cytokinesis the cytoplasm divides by the formation of a cleavage furrow, which pinches the cell in two producing two identical daughter cells. Meiosis Meiosis is cell division to produce the gametes or sex cells. It is similar to mitosis but chromosome replication is followed by two consecutive cell divisions resulting in four daughter cells with half complements of chromosomes. Like mitosis, meiosis is preceded by chromosome replication in interphase I. 1. Interphase I: Chromosome replication as for mitosis. 2. Prophase I: The chromosomes condense and are individually visible. The difference here is homologous chromosomes, each made up of two chromatids, come together in pairs to form a complex of four chromatids or tetrads. These chromatids are in close proximity and cross-over in numerous places along their length. 3. Metaphase I: The tetrads align in the centre of the meiotic spindle. 4. Anaphase I: The spindle apparatus separates the chromosomes and pulls the homologous chromosomes to opposite ends of the cell. The homologous chromosomes separate from one another but the sister chromatids are still joined at their centromere. For example, both copies of chromosome 12 in the first cell are copied making four chromatids. The chromatids derived from the father’s chromosome 12 are joined at the centromere as is the case for the mother derived chromosome 12. Each one of these sits on either side of the metaphase plate and get separated in anaphase I. 5. Telophase I and Cytokinesis: Nuclear envelopes form around the chromosomes at either end of the cell and the cell divides. 6. Prophase II: In each of the daughter cells a spindle apparatus appears and the nuclear envelope fragments. 7. Metaphase II: The chromosomes align individually along the metaphase plate, this time as in mitosis, not with homologous chromosomes side by side. 8. Anaphase II: The centromeres separate pulling sister chromatids apart and toward each end of the cell. 9. Telophase II and Cytokinesis: Nuclei form at each end of the cell, the cytoplasm divides producing four daughter cells each with a haploid number of chromosomes. 2. Define autosomal recessive inheritance Autosomal recessive is a mode of inheritance of genetic traits located on the autosomes. In opposition to an autosomal dominant trait, an autosomal recessive trait only becomes phenotypically apparent when two similar alleles of a gene are present. In other words, the subject is homozygous for the trait. Recessive genetic disorders occur when both parents are carriers and each contributes an allele to the embryo , meaning these are not dominant genes. As both parents are heterozygous for the disorder, the chance of two disease alleles landing in one of their offspring is 25%. 50% of the children (or 2/3 of the remaining ones) are carriers. When one of the parents is homozygous, the trait will only show in his/her offspring if the other parent is also a carrier. In that case, the chance of disease in the offspring is 50%. 3. Explain dominant and recessive mutations at the molecular level A dominant mutation encodes a gain of protein function which results in increased protein production. This causes a gain in function (may not be what it is meant to do normally) If you have one it will alter the phenotype. A dominant mutation is apparent in both the homozygous dominant and heterozygous states. A recessive mutation encodes a loss of protein function. E.g. Due to deletion of the gene encoding the protein. A recessive mutation is only seen in the homozygous recessive state. No (or minimal) effects are seen in the heterozygous state (carrier) because protein production from the normal gene generally makes up for the deficiency. Which is why carriers are always carry recessive diseases, as the recessive mutation is covered up by the normal gene and the disease is not expressed. 4. Explain how uniparental disomy may occur Uniparental disomy Occasionally, autosomal recessive disorders can arise through a mechanism called uniparental disomy, in which a child inherits two copies of a particular chromosome from one parent and none from the other. If the chromosome inherited in this uniparental fashion carries an autosomal recessive gene mutation, then the child will be an affected homozygote. Recurrence risk for future siblings is extremely low. 5. Define linkage and explain how mapping can be deduced from linkage analysis Linked genes are genes that tend to be inherited together in genetic crosses because they are located close together on chromosomes as a unit. I’ll explain from the beginning; skip ahead if need be. During Meitoic Interphase (at the very beginning of Meiosis) the chromosomes duplicate (from 46 to 92) and the two chromosomes form a sister chromatid, this is still considered as a chromosome even though it has two sets of DNA (thus there are still 46 chromosomes). But now the structure is referred to as a homologous chromosome. During Prophase I in Meiosis the homologous chromosomes (maternal and paternal) find each other. These homologous chromosomes originally came from the mother and father of the individual. Thus, maternal and paternal chromosomes, as homologs, line up side by side. (Ie maternal homologous chromosome 7 with paternal homologous chromosome 7). The homologous chromosomes are not identical: they have different combinations of alleles at the thousands of loci along each chromosome. The homologous chromosomes then pair up. The 4 chromatids of the two homologs actually zip together, and the chromosomes exchange parts of their arms. Enzymes cut the chromosomes into pieces and seal the newly combined strands back together in an action called crossing-over or recombination. Because of the way genes are situated along chromosomes, genes that are very close together spatially are more likely to be inherited together. When genes are so close that they are inherited together (all or some of the time) then they are considered to be linked. The process of linkage is really a determination of how often recombination occurs between two or more traits. If two genes are close enough together they will end up being linked more than 50% of the time. For example the two genes that code for blond hair and blue eyes may be very close together and are thus linked. They will there for be inherited together. In another example: a disease state is associated with blue eyes, the gene for the disease and the gene for blue eyes are linked, and are probably in close proximity on the chromosome. If the genes are further apart on the chromosome, there is more chance of recombination: they will act independently of each other. Knowledge of gene linkage is useful in genetic mapping. A genetic map represents the linear sequence of genes along a chromosome. The further apart two genes are on a chromosome, the more likely it is that crossing over will occur between them and therefore the more likely it is that recombinant individuals will occur. Crossover data can be used to construct a genetic map or linkage analysis. So if a large percentage of recombinants for two linked genes is seen in a population, you would assume those genes were far apart on the chromosome. (Brown eyes and big feet) Conversely if there were few recombinant individuals, you’d assume the linked genes were fairly close together. (Blond hair and blue eyes) The distance between genes is measured in centimorgans, which refers to the probability of recombination. Each % is a cetimorgan so a 20% recombination frequency is 20 centimorgans. Based on this theory, recombination frequencies are used to determine the relative positions of different genes on the chromosome. Chromatid: One half of a replicated chromosome Chromosome: Linear or circular strand of DNA Homologous Chromosomes: Two chromosomes that are identical in shape and structure and carry the same genes Homolog: Homologous Chromosome