PLNT_CELLrevWang
advertisement

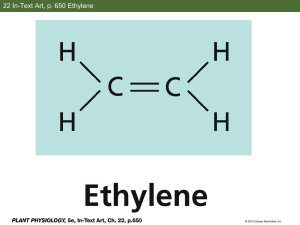
Dear Dr. Patterson: I apologize for the delay in contacting you about your manuscript. We have now received all reviews of your manuscript entitled "Identification of Important Regions for Ethylene Binding and Signaling in the Transmembrane Domain of the ETR1 Ethylene Receptor of Arabidopsis," which you may view by visiting http://submit.plantcell.org. The work you describe is very interesting, but your manuscript requires some revision. We will consider it appropriate for publication if you return an acceptably revised version that is fully responsive to our concerns and questions. I had many of the same comment/questions as Reviewer 2 (mislabeled Figure 4, missing controls and the need for some statistical analyses). In addition, would you please comment on the following in your response to me. What is the basis for concluding that the P110A mutation does not significantly alter the ethylene-binding ability of ETR1 (Figure 2)? What is your explanation for the effects of altering residues (His and Cys in particular) that are not typically found in transmembrane segments and are likely to be copper coordinating residues relative to the model for ethylene binding involving conjugation of the double bond of ethylene to a bound copper? Please note that manuscripts will not be considered further that do not comply with all journal requirements described in the Instructions to Authors. When you have had an opportunity to study the reviews, please contact The Plant Cell editorial office if you believe you will need more than 30 days to submit a revision. If a revision is not returned within 30 days and you have not been granted an extension, the manuscript may be declined and treated as new submission. When uploading the revised version, please be sure to use the Word highlight function to indicate the changes in the text that you have made in response to the coeditor and reviewer recommendations. Include an itemized list of all changes made in response to each of the reviewer's suggestions in the "Response to Reviewers" section; please note that reviewers do not have access to your cover letter and so will not be able to read your itemized list if it is loaded there. The Plant Cell strongly encourages authors to deposit novel materials such as genetic stocks in the appropriate genetic stock center, e.g., ABRC/NASC for Arabidopsis stocks, so that they will be available conveniently to the entire community. Corresponding authors who are ASPB members at the time their paper is accepted receive a $20 discount per page on article page charges. If you are not already a member, we encourage you to join. Please visit http://www.aspb.org/memberjoin?mktgsource=PCPRESS. If you have any questions about these revisions or the online submission procedures, please feel free to contact me or the Manuscript Manager, Annette Kessler, respectively. We look forward to receiving your revised manuscript. Sincerely, Karen Schumaker Coeditor, The Plant Cell PLANTCELL/2006/044537 Identification of Important Regions for Ethylene Binding and signaling in the Transmembrane Domain of the ETR1 Ethylene Receptor of Arabidopsis Wuyi Wang, Jeff J Esch, Shin-han Shiu, Hasi Agula, Brad M Binder, Caren Chang, Sara E Patterson, and Anthony B Bleecker Date Received: 6 Jun 2006 Article Types: Research Article TOC Category: Research Article Corresponding Author: Sara E Patterson Subject Areas: hormonal regulation; hormone biology; signal transduction Supplemental Files: 0 Reviewer 1 Review Comments Reviewer 2 Review Comments Reviewer 3 Review Comments Reviewer 1 Review Comments... The ethylene receptors are known to function as negative regulators of the relatively well-understood ethylene signaling pathway. Among the different structural features present in these receptors, the hydrophobic amino terminus formed by three to four membranespanning domains is responsible for the interaction between the receptor and ethylene and is referred to as the ethylene-binding domain (EBD). The ethylene binding properties of the EBD can be studied in yeast expressing the corresponding EBDs. Interestingly, proteins with sequence similarity to the ethylene receptors can be found in non-plant species, and a protein from Synechocystis with sequence similarity to ETR1 has been shown to bind ethylene. In this manuscript, the authors address two interesting questions: 1) What is the evolutionary origin of the ethylene receptors? 2) What is the structure-function relationship of the EBD? To address the first question, the authors examined the ethylene binding properties of an extensive variety of species from all kingdoms. Representatives of land plant species including ferns and bryophytes showed ethylene binding. Among non-land plant species, only the algae Chara and cyanobacteria from the Synechocystis clade were found to possess ethylene binding activity. These results point to a plastidic origin of the ethylene receptors. It would be helpful if in figure 1 the authors could display a tree showing the evolutionary relationship between the species studied. An extensive search for putative EBD sequences was carried out. Sequences related to the EBD were discovered not only in cyanobacteria, but also in other diverse species of bacteria. Again, it would be helpful to have an evolutionary tree of these species to make it easier for the readers to follow this part of the work. To better understand the relationship between structure and function, 41 highly conserved aa of ETR1 were mutated and the binding activity of the corresponding EBD in yeast, as well as the in planta signaling activity of the mutant EBD forms, were examined. In this study, specific residues in helixes I and II were found to be critical for ethylene biding, whereas residues in helix III generally had only minor effects. In addition to the mutations that abolish ethylene binding and confer constitutive ethylene insensitivity, several other mutations were found that also result in ethylene insensitivity but do not affect the binding of ethylene. These results suggest that the aa clustered in helix III and bottom of helix I are critical for transmitting the signal generated upon ethylene binding. In summary, this is a comprehensive study that provides new information on the role of specific residues in the EBD of the receptors. By combining sequence information with the results of ethylene-binding studies and in planta functional assays, new insights into the relationship between structure and function of the ethylene receptors have been obtained. Furthermore, the aforementioned survey of the ethylene binding capacities of different plant species suggests plastidic origin of the receptors. Reviewer 2 Review Comments... In the first part of this manuscript, Wang et al. describe the identification of ethylene receptor-like sequences in species of different kingdoms and the use of a bioinformatics approach to get insight into the potential origin and distribution of ethylene receptors. The bioinformatics data are well done and show that ethylene-binding domain (EBD) containing sequences are found largely in land plants, Chara and a group of cyanobacteria. The bioinformatics approach is accompanied by ethylene binding assays using crude material from selected organisms. In the second part of the manuscript, the authors describe their use of analysis to identify additional amino acid residues in the EBD of ETR1 which might be crucial for ethylene binding and/or signal transmission to the cytoplasmic transmitter domain of the ethylene receptor. The overall impression of the manuscript is very positive. However, I have some criticisms and points the authors should address: Major comments: • The authors mention, without showing the available data, that “all of the ETR1 mutants formed homodimers in yeast and could be converted into monomers by treatment with DTT, indicating that the mutations did not disturb the native structure of the receptor and that the ETR1 mutants were still membrane-bound proteins” (page 8/9). As these data are so crucial for the interpretation of functional results later on, I would recommend including them as a supplemental figure, which should also contain a short description of the method. • For the same reason the authors should present the immunoblot results of ethylene receptor expression, which they used for protein quantification (!), in Figure 1. • Regarding the hypocotyl length determination, there are no statistics given (e.g. how often was the experiment repeated, how many seedlings were measured, SD values). • A major drawback of the manuscript is the missing protein expression data for the transgenic lines expressing the different ETR1 mutants. I would like to see immunoblot data for at least some representative lines of each response group. • The authors should explain why they switched to the ers12 etr1-7 double mutant for the investigation of the S98A and P110A EBD mutants. • Figure 4B does not show rosette size data as mentioned in the text but results of hypocotyl length measurements. Please solve this problem. Minor comments: • The paper “Chang and Mount, 2002” is not mentioned in the REFERENCES • The authors write that the etr1-6 etr2-3 ein4-4 mutant has a constitutive ethylene-response phenotype. However, in Figure 3, they show that this mutant is still responsive to ethylene (air: 9 mm; ethylene: 3 mm)! Please comment on this. • For non-specialist readers, please give a substantiation of the AVG experiment and a short interpretation of the results. • Please explain in more detail why a “large number of mutations that cause a drastic….ethylene binding” allows the suggestion “that the transduction of……requires only subtle changes in steric structure”. • The last paragraph is very difficult for non-specialist readers to understand. As the introduction of a possible intermediate state (state II) of ethylene receptors is crucial for the understanding of receptor function and the mutant phenotypes, an in-depth discussion should be provided. Reviewer 3 Review Comments... The pathway of hormone signal transduction for ethylene has been advanced through genetic studies identifying multiple components of ethylene signaling including a family of receptor molecules. While numerous steps in the pathway have been identified and ordered based on epistatic interactions, the biochemistry of ethylene signaling remains largely unknown. Here the authors provide a comprehensive mutational analysis on the ethylene binding domain (EBD) of the ETR1 EBD and assay ethylene binding capacity of a large collection of EBDs derived from genes representing diverse taxa. This latter activity was based on an informatics assessment of EBD prevalence and yielded results consistent with the prior hypothesis that the origin of plant ethylene receptors traces back to symbiotic acquisition of a cyanobacterial genome. The mutational analysis clarifies the borders of the EBD, confirms helicies I and II as the basis of the ethylene binding pocket and has facilitated development of a more in depth model of receptor function and functional states. The manuscript is well written and while the amount of work is considerable, the experimental analysis is relatively straight-forward and based on systems that are well established in the lab. As such it is not surprising that the data is clear and of high quality. The manuscript itself is very well written and a tribute to the former mentor of the group. The only point I would ask the authors to address is state II of their model. I am not quite sure I follow the logic that uses the fact of mutations uncoupling receptor/ligand binding and receptor pathway suppression to infer the existence of such a state in normal biochemical activity of receptors. I may be missing something here but it seems that states I and III are clear and II appears a bit forced. The authors should clarify their logic in the Discussion.