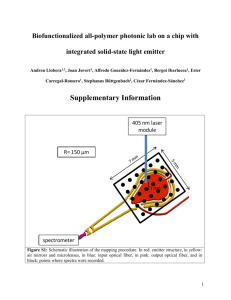
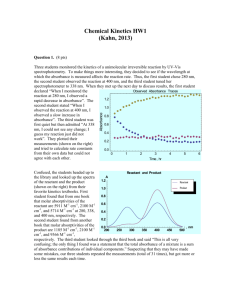
bit_23038_sm_suppInfo
advertisement
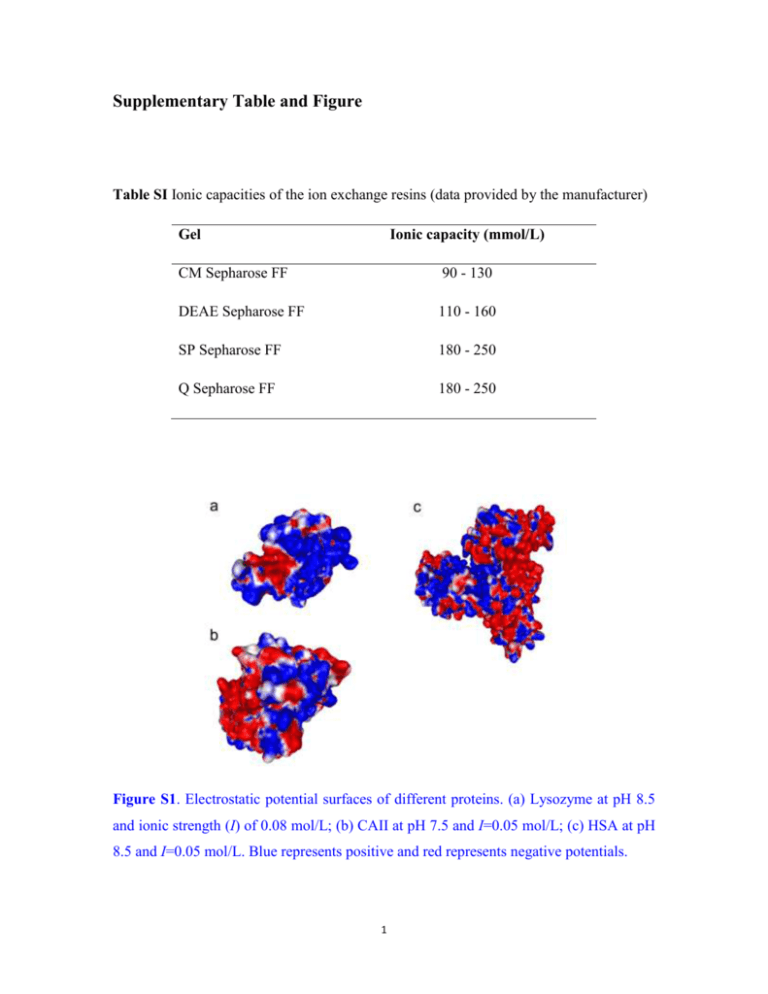
Supplementary Table and Figure Table SI Ionic capacities of the ion exchange resins (data provided by the manufacturer) Gel Ionic capacity (mmol/L) CM Sepharose FF 90 - 130 DEAE Sepharose FF 110 - 160 SP Sepharose FF 180 - 250 Q Sepharose FF 180 - 250 Figure S1. Electrostatic potential surfaces of different proteins. (a) Lysozyme at pH 8.5 and ionic strength (I) of 0.08 mol/L; (b) CAII at pH 7.5 and I=0.05 mol/L; (c) HSA at pH 8.5 and I=0.05 mol/L. Blue represents positive and red represents negative potentials. 1 Supplementary Methods Protein Assay Lysozyme activity was assayed using Micrococcus lysodeikticus as the substrate (Roux et al., 1997). In the assay, 1.30 mL of 0.25 mg/mL substrate solution was mixed completely with 0.10 mL protein solution at 25 oC. The decrease in the absorbance of the mixture at 450 nm was then recorded. The slope of the absorbance curve of the first 90 s was used for calculation of the enzyme activity. RNase A activity was assayed by the addition of the aliquot to a 450 μM cCMP solution (0.1 M Tris–HCl, 1 mM EDTA, pH 6.0) at 25 oC (Beld et al., 2008). The increase in the absorbance of the mixture at 284 nm was then recorded. The slope of the absorbance curve of the first 90 s was used for calculation of the enzyme activity. Esterase activity of carbonic anhydrase was assayed using p-nitrophenyl acetate (pNPA) as the substrate as described previously (Baynes et al., 2005). Briefly, 15 μL sample was added to 1 mL of 0.05 M Tris-HCl, 5 mM EDTA, pH 7.5, and 10 μL of 100 mM pNPA in acetonitrile. The kinetics of hydrolysis of pNPA was observed by the increase in absorbance at 400 nm. Hydrolysis rates were corrected for the hydrolysis of pNPA by the buffer. Hydrolysis rates were converted to concentration of active protein via a standard curve constructed from dilutions of known concentrations of the native protein. For BSA, its recovery was measured by reverse phase high-performance liquid chromatography (RP-HPLC) according to Langenhof et al. (2005) with a little modification. RP-HPLC analysis was performed on an Agilent 1100 Series HPLC-system. Samples containing 0 to 10 μg protein were injected onto Zorbax Eclipse XDB-C8 column (4.6 × 150 mm; Agilent Technologies). A binary gradient using solvent systems 2 A (0.1% TFA/water) and B (0.1% TFA/acetonitrile) was run at a flow rate of 1 mL/min. After a 5-min equilibration period at 37% B, a linear gradient from 37% to 38% B in 2 min was implemented. A second gradient from 38% to 58% was implemented after it was held at 38% for 10 min. Protein elution was performed at room temperature and monitored by measuring absorbance at 214 nm. The mass of total protein eluted from the RP-HPLC column was quantitatively determined by peak integration, based on standard curves obtained from calibration using native and denatured BSA standards. References Baynes BM, Wang DIC, Trout BL. 2005. Role of arginine in the stabilization of proteins against aggregation. Biochemistry 44(12):4919-4925. Beld J, Woycechowsky KJ, Hilvert D. 2008. Catalysis of oxidative protein folding by small-molecule diselenides. Biochemistry 47(27):6985-6987. Langenhof M, Leong SSJ, Pattenden LK, Middelberg APJ. 2005. Controlled oxidative protein refolding using an ion-exchange column. J. Chromatogr. A 1069(2):195201. Roux P, Delepierre M, Goldberg ME, Chaffotte A-F. 1997. Kinetics of secondary structure recovery during the refolding of reduced hen egg white lysozyme. J. Biol. Chem. 272(40):24843-24849. 3