Procedure

Molecular Cell Biology Laboratory
Instructor: Elmar Schmid, Ph.D.
Setting up of a restriction digest
Generally, restriction enzyme cleavage is a relatively simple procedure, accomplished by incubating the (plasmid or PCR-derived) DNA in the presence of the selected restriction enzyme(s) in the appropriate “digestion buffer’
Necessary Equipment:
Water bath (adjusted to 37 o C)
Microcentrifuge
Vortex
Adjustable-volume digital pipettes (e.g. Eppendorf, Finnipette)
Pipette pump
1 Plastic rack (for 1.5ml reaction tubes)
Ice bucket (filled with crushed ice)
Required Materials:
- Sterile, DNAse-free water (= ddH
2
O) (50ml; Epicentre, Cat# W7350ML)
- Purified PinPoint Xa-1T vector
- 10x TA Buffer (1.5ml; Epicentre; Cat# TA6115)
- 2 selected restriction enzymes.
cutting upstream and downstream of MCS region
e.g. Bam H I and Sma I (see your vector map)
store at -20 o C and keep on ice during the experiment
- 6x gel loading buffer
- Sterile plastic reaction tubes (polypropylene, 1.5ml)
- Pipette tips (yellow, blue)
Procedure: a. Take all necessary restriction enzymes and the TA restriction buffer out of the freezer and place them on ice in an ice bucket
the enzymes are in a glycerol buffer and do not need to be
thawed; but make sure they are not solid frozen!
thaw the 10x TA buffer in your hands then place on ice b. For each planned plasmid DNA digestion (in your case: for each plasmid DNA isolated from the positive bacterial clones: = 10) place the same number (= 10) of sterile 1.5ml reaction tubes in a plastic rack and label them; Place two additional reaction tubes (labeled #11 and #12) into the rack and use these ones for important control reactions
make sure you use a waterresistant marker so you won’t loose
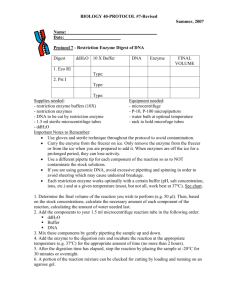
your info during the following incubation in the water bath c. Pipette the following reagents into the prepared reaction tubes in your rack
make sure you pipette the reagents in the sequence from top to
bottom (see Table below)
1
Molecular Cell Biology Laboratory
Instructor: Elmar Schmid, Ph.D.
Pipetting Scheme:
Reagent ddH
2
O
Restriction Buffer
DNA
(e.g. PCR amplicon or Vector DNA)
Restriction enzyme 1
Restriction enzyme 2
Conc./
Amount
10x
1 -
5μg/50μl
1
– 5 units
1 – 5 units
Volume
[ μl ]
13 – x – y
2
5 x y
Total: 20 d. Close the lids and gently mix the contents by gently flicking the tubes with your fingers
make sure the mix remains at the bottom of the tube afterwards
otherwise, briefly spin the sample(s) in a microcentrifuge e. Insert the tubes in a Styrofoam float, place them into the water bath (adjusted to 37 o C) and incubate the samples for 1 hour without shaking
generally: 1 U restriction enzyme digests 1μg of DNA in 60min under
optimum assay/buffer conditions! f. After 1
– 1.5 hrs., take the samples out of the water bath and pipette 4μl of the
6x gel loading buffer to each of the samples g. Briefly vortex all samples and load them into the pockets of a previously prepared agarose gel (for preparation: see previous lab section below) h. Perform agarose gel electrophoresis to separate the DNA fragments of your restriction digest samples and visualize the DNA with the help of an UV transilluminator (for procedure: see agarose gel electrophoresis lab manual) i. Document your results on the gel using the Polaroid gel documentation system and discuss the outcome with your group and your instructor
2
Molecular Cell Biology Laboratory
Instructor: Elmar Schmid, Ph.D.
Assignment:
____________________________________
(Student Name)
1. Draw or paste the gel image showing the results of your restriction digest of the
PinPoint vector purified from your positive clones in to the available space (box) below.
- label the individual lanes and annotate the molecular weights of the: a. DNA standard/marker b. Restriction fragments of the purified plasmid DNA of your different JM109 clones
Agarose gel electrophoresis result of the restriction digests
MW
[kDa]
Lanes
2. Write down the incubation protocol for your chosen control reactions (reaction tubes
#11, 12) during the restriction digest experiment into the table below.
[ μl ]
Reagent Tube #11 Tube #12 ddH
2
O
TA Buffer
PinPoint Xa-1T vector
Restriction enzyme 1
Restriction enzyme 2
3