HIV Virology Handout
advertisement

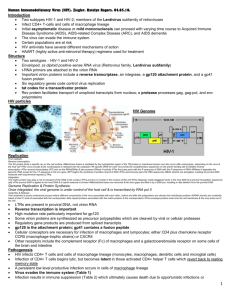
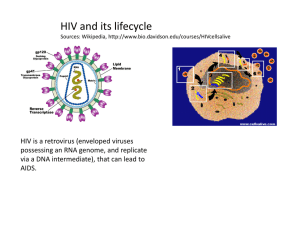
Lecture: Retrovirus Biology Michael Para, M.D. Professor of Medicine Division of Infectious Diseases N-1143 Doan Hall Phone: 614-293-5666 michael.para@osumc.edu Learning objectives 1. Recognize the unique features and interrelationship of simple and complex retroviriuses 2. List the major structural components of HIV 3. Describe how HIV replicates 4. Know mechanisms of action for antiretroviral drug classes 5. Recognize how HIV becomes drug resistant and how this is prevented Suggested reading 1. Review Articles Medical Progress: HIV Drug Resistance N Engl J Med 2004; 350:1023-1035, Mar 4, 2004 Retroviridae Definition 1. *Single stranded + sense RNA virus, with 5’cap, poly A tail (like mRNA),*2 identical RNA strands in capsid structure in lipid envelope (see next page); they replicate through liner double stranded DNA intermediate using reverse transcriptase enzyme (RT), and can integrate into host cell DNA 2. *All have ≥ 4 genes- gag (core proteins e.g capsid), pol (RT, IN integrase), pro (protease), env (envelope) a. Simple retroviruses have just these 4 genes (see A in Figure 1) b. Complex retroviruses have spliced regulatory genes controlling protein synthesis & replication (see B in Figure below) 3. Long terminal repeat sequences (LTR) flank all retroviral genome a. LTR contain all signals necessary for gene expression- enhancers, promoters, transcription initiation and transcription termination sequences and polyadenylation signal. Host transcription factors bind to LTR b. *Integrated retrovirus (= provirus) has two LTRs; the 5' LTR acts as RNA polymerase II promoter whereas the 3' LTR functions as the terminator sequence. Figure 1 simple retrovirus Complex retrovirus LTR a. Oncoviruses = transforming retroviruses, usually do not kill cell, but alter cell’s growth, can immortalize cell a. Animal - both chronic (no onc gene) and acute (with onc) as murine or avian leukemia viruses b. Human-Human T Lymphotropic Virus I and II (HTLV-I and HTLV-II) b. Lentiviruses = non-transforming, cytopathic (lyse cells) retroviruses, can have long latency but when replicating leads to viremia, cell death and typically causes neurologic or immunologic disease a. Animal STLV-III (SIV) – causes no illness in natural host but causes simian AIDS in other primate species b. HIV = Human Immunodeficiency Virus, more complex a. formerly HTLV-III (3rd human retrovirus) now HIV-1 c. HIV-2, 2nd human retrovirus also associated with acquired immunodeficiency disorder (AIDS) in west Africa Structure of HIV 1. *Enveloped single stranded RNA virus 9750 nucleotides in length, 100nm in size a. *lipid bilayer envelope with dense cylindrical core and external spikes, 2 RNA genomes per virion b. major surface antigen is glycoprotein= gp120, transmembrane glycoprotein = gp41 2. *Structural genes a. env gene–major surface antigens, 2 glycoproteins;gp 41 & gp120 i. gp120 - responsible for binding to CD4, and co-receptor ii. gp41 - transmembrane protein, leads to fusion of cell membrane and viral envelope i. gag gene - 4 internal structural proteins cleaved from polyprotein precursor – nucleocapsid core, matrix protein on inner surface of envelope and 2 nucleic acid binding proteins b. pol gene - includes 3 enzymes - all drug targets i. *reverse transcriptase (RT) –RNA dependent DNA polymerase and *ribonuclease activity (66kD), RT is site of activity for nucleoside analogue anti-retroviral drugs (below) ii. *protease - cleaves the polyprotein made from the gag and pol genes, it is site of activity for protease inhibitor anti-HIV drugs iii. integrase - integrates viral DNA into host cell, site of activity for integrase inhibitor anti-retroviral drugs Reverse transcriptase Protease with inhibitor 3. *Non-structural & Regulatory Genes - protein products interact with responsive elements on proviral DNA are produced by noncontiguous genes --> spliced mRNA (see figure below), these regulate HIV replication TAT, REV, NEF, VPU, VPR, VIF Replication of HIV Watch www.youtube.com/watch?v=RO8MP3wMvqg / or www.hhmi.org/biointeractive/media/hiv_life_cycle-lg.mov 1. Binding of virus to CD4 cell membrane, membrane fusion & penetration (more explanation during Immunology of HIV lecture) a. gp120 binds with high affinity to CD4 molecule, inducing conformational change in gp120 shape b. conformational change in gp120 exposes new gp120 viral surface that bind to a 2nd protein called the “co-receptor” c. coreceptor in T-cells is CXCR4, viruses using CXCR4 co-receptor are call T cell or lymphotropic viruses d. coreceptor on macrophages, is CCR5, viruses using CCR5 coreceptor are called macrophage tropic viruses e. CCR5 & CXCR4 are chemokine receptors, which function as part of normal immune response 2. Fusion a. The binding of gp120 to coreceptor exposes gp41 hydrophobic tail that pierces into cell's membrane b. gp41 folds on itself pulling virus into cell, initiating cell - virus fusion 3. *Nucleic acid synthesis a. RNA genome cDNA by reverse transcriptase(RT), ribonuclease function on RT digests HIV RNA, leaving only HIV cDNA b. cDNA copied by RT dsDNA circularizes using matching LTR c. The “preintegration complex” of dsDNA + integrase enters nucleus d. dsDNA is incorporated into host cell chromosome (proviral DNA) using viral integrase e. From proviral DNA cellular DNA polymerase can make various new mRNA, some spliced for viral proteins, and copies of genomic RNA f. genomic HIV RNA and mRNA migrate into cytoplasm long polyproteins made and then cleaved by viral protease making specific proteins assemble new virus bud off of cell membrane Regulation of Viral Replication– replication depends on cell’s interaction with LTR 1. HIV replication from host proviral DNA restricted until T cell activated a. Some HIV carrying cells may not express any viral mRNA (virus exists in quiescent latent state) b. Activation of host cell induces cell transcription factors that activate latent HIV via LTR interaction c. Balanced viral protein synthesis can allow some low level viral replication without cell death *Antiretroviral agents- inhibit enzymes needed for replication or block viral entry 1. *Reverse Transcriptase (RT) Inhibitors – block new synthesis of complete viral RNA/DNA strands a. *Nuceloside analogues– drugs are incorporated into elongating viral DNA/RNA causes chain termination b. Non-Nuceloside analogues – interfere with binding of viral RNA to viral RT 2. *Protease Inhibitors (PI)– stops viral protease from cleaving viral polyproteins preventing viral maturation 3. Integrase inhibitors –blocking viral DNA integration into cellular DNA 4. Entry inhibitors a. Attachment inhibitors – receptor antagonist e.g. antibodies i. Co-receptor antagonists – maraviroc interferes with gp120 binding to CCR5 co-receptor on CD4 b. Fusion inhibitors - T-20 binds to gp41 blocking gp41 folding on itself which prevent viral envelope and cell membrane fusion *Viral Heterogeneity - 1-25% variation in genome from one HIV-1 isolate to next 1. RT has high rate of transcriptional errors, about 1 to 5 per 10,000 bp, RT has no "correcting" function 2. This mutational mechanism allows HIV to evolve and optimize growth/persistence in given host 3. Mutations may alter growth rates or antigenicity, or protein function a. Marked heterogeneity seen in env gene b. These can lower neutralization by antibodies, c. Can alter drug biding site making virus drug resistant d. "mutant" virus can escape host defenses or antivirals, then multiply e. *Strains with drug resistance emerge in presence of antiviral meds *Antiretroviral drug resistance and effective therapy 1. Monotherapy with any agent invariably fails due to mutation and subsequent drug resistance 2. Combination therapy- treatment “cocktails”- e.g. triple drug regimens suppress replication 1. With replication suppressed, no new mutations so viral evolution slows/stops, and development of drug resistance from mutations in viral enzymes does not occur. Fully suppressive therapy remains effective