Appendix 1: Glossary accessory genome The collection of genes
advertisement
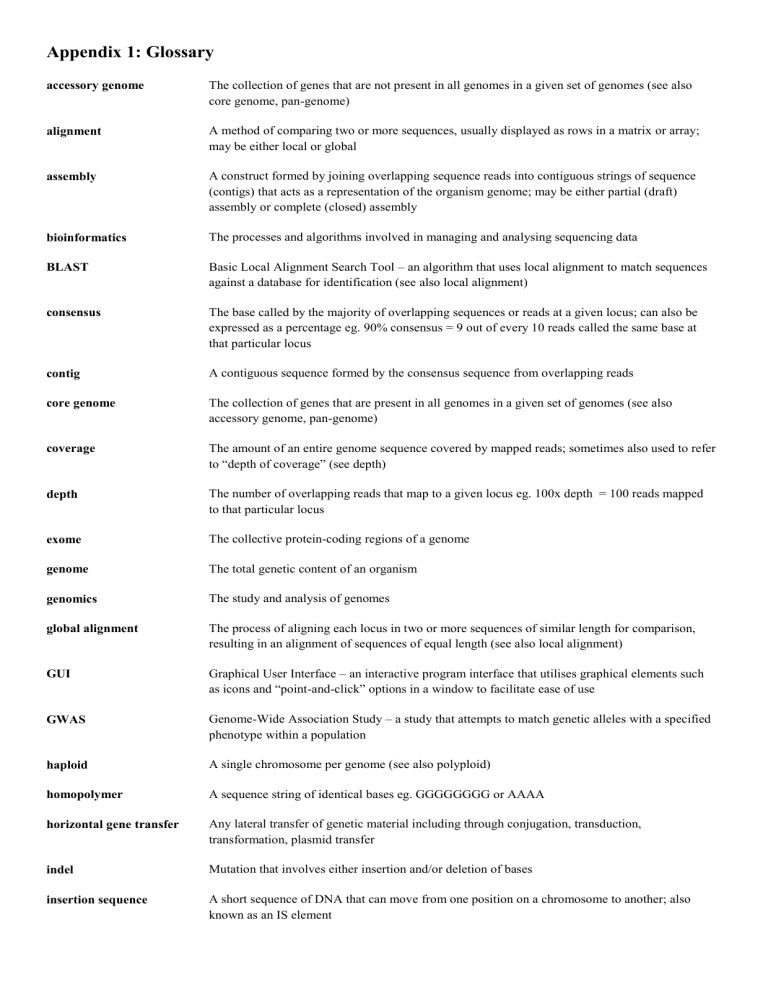
Appendix 1: Glossary accessory genome The collection of genes that are not present in all genomes in a given set of genomes (see also core genome, pan-genome) alignment A method of comparing two or more sequences, usually displayed as rows in a matrix or array; may be either local or global assembly A construct formed by joining overlapping sequence reads into contiguous strings of sequence (contigs) that acts as a representation of the organism genome; may be either partial (draft) assembly or complete (closed) assembly bioinformatics The processes and algorithms involved in managing and analysing sequencing data BLAST Basic Local Alignment Search Tool – an algorithm that uses local alignment to match sequences against a database for identification (see also local alignment) consensus The base called by the majority of overlapping sequences or reads at a given locus; can also be expressed as a percentage eg. 90% consensus = 9 out of every 10 reads called the same base at that particular locus contig A contiguous sequence formed by the consensus sequence from overlapping reads core genome The collection of genes that are present in all genomes in a given set of genomes (see also accessory genome, pan-genome) coverage The amount of an entire genome sequence covered by mapped reads; sometimes also used to refer to “depth of coverage” (see depth) depth The number of overlapping reads that map to a given locus eg. 100x depth = 100 reads mapped to that particular locus exome The collective protein-coding regions of a genome genome The total genetic content of an organism genomics The study and analysis of genomes global alignment The process of aligning each locus in two or more sequences of similar length for comparison, resulting in an alignment of sequences of equal length (see also local alignment) GUI Graphical User Interface – an interactive program interface that utilises graphical elements such as icons and “point-and-click” options in a window to facilitate ease of use GWAS Genome-Wide Association Study – a study that attempts to match genetic alleles with a specified phenotype within a population haploid A single chromosome per genome (see also polyploid) homopolymer A sequence string of identical bases eg. GGGGGGGG or AAAA horizontal gene transfer Any lateral transfer of genetic material including through conjugation, transduction, transformation, plasmid transfer indel Mutation that involves either insertion and/or deletion of bases insertion sequence A short sequence of DNA that can move from one position on a chromosome to another; also known as an IS element local alignment The process of aligning each locus in a shorter sequence against a corresponding section of a longer sequence or database for comparison (see also global alignment, BLAST) locus A position in a genome sequence occupied by a single nucleotide metagenomics The study and analysis of all the genomic content in a sample (frequently involves multiple organisms) models of DNA evolution Models that aim to predict the rate and direction of nucleotide substitution during evolution eg. Jukes & Cantor, Generalised Time-Reversible, Hasegawa, Kishino & Yano; usually incorporated into maximum likelihood and Bayesian models to estimate a phylogeny molecular clock The natural background rate of evolutionary change in a genome that is assumed to exist N50 A genome assembly metric – the N50 is the contig length where the sum of the contigs of that length or longer comprise ≥50% of the total sum of contig lengths, and the sum of the contig of that length or shorter comprise ≥50% of the total sum of contig lengths neighbour-joining A rapid clustering method to draw a phylogenetic tree; other more probabilistic methods include maximum likelihood and Bayesian analysis open source Software that is publicly available and free pan-genome The total collection of genes present in a given set of genomes. Pan-genome = core genome + accessory genome Phred-score A per-base score to indicate the quality and accuracy of sequencing; Q10 = 1 error per 10 bases, Q20 = 1 error per 100 bases, Q30 = 1 error per 1000 bases phylogeny The history of the genetic evolution of a group of organisms; usually represented by a phylogenetic tree, where branches represent the genetic differences between organisms polyploid Multiple chromosomes per genome (see also haploid) read clipping A post-sequencing bioinformatics quality-control process where raw sequence reads are filtered and trimmed to remove indexing adaptors attached to the DNA sequence during library preparation, and to remove low quality sequence regions read mapping The process of aligning/matching sequence reads against corresponding regions of a reference genome reads The raw, usually overlapping, unanalysed sequence output from a genome sequencer, that can be either mapped to a reference genome, or assembled de novo into contigs; length varies depending on the sequencer and intended study recombination Any process involving the horizontal exchange of genes between two organisms reference genome A representative (usually complete / closed) genome scaffold A genome framework, comprising several contigs, separated by expected gaps of a known size SNP Single nucleotide polymorphism – a single base variation transcriptome The sum of all transcribed RNA produced by a set of organisms








