Supplementary Information (doc 54K)
advertisement

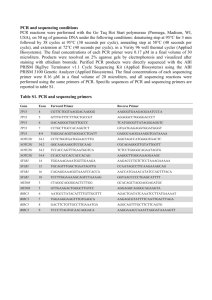
Supplementary Table 1. Oligonucleotide primers for RT-PCR to sequence Exon 34 of the Notch-1 PEST domain of the MT-1 cell line Notch1 Nucleotide Primer Gene Oligo Sequence Sequence Application (NM_017617.3) Notch1, Nucleotides 60615’-GTCAACGCCGTAGATGACCT-3’ N692 Exon34 RT-PCR, Sequencing 6080 Notch1, Nucleotides 70935’-AGCTCATCATCTGGGACAGG-3’ N703 Exon34 RT-PCR, Sequencing 7075 Notch1, Nucleotides 75055’-CTTACAGATGCAGCAGCAGAACC-3’ N714 Exon34 RT-PCR, Sequencing 7528 Notch1, Nucleotides 86665’-TGTGTTGCTGGAGCATCTTC-3’ N704 Exon34 Notch1, N696 RT-PCR, Sequencing 8647 Nucleotides 6809- 5'-CACCTCGTCTCTCCCACCT-3' Exon34 Notch1, N716 RT-PCR, Sequencing 6827 Nucleotides 8088- 5'-CCTGGCATCCACAGAGCGCAC-3' Exon34 Notch1, N695 RT-PCR, Sequencing 8068 Nucleotides 6338- Internal Primer, 6357 Sequencing Nucleotides 7293- Internal Primer, 7312 Sequencing Nucleotides 7745- Internal Primer, 7764 Sequencing Nucleotides 8331- Internal Primer, 8310 Sequencing 5'-TGGACGAGTACAACCTGGTG-3' Exon34 Notch1, N697 5'-GAGCTTCCTGAGTGGAGAGC-3' Exon34 Notch1, N698 5'-CGACCAGAGGAGCCTTTTTA-3' Exon34 Notch1, N715 5'-ACAAGCATGCTTGCAAGAAACC-3' Exon34 1 Supplementary Figure 1 Ex vivo triple combination treatment did not induce significant apoptosis of PBMCs from normal donors. (A)PBMCs from normal donors were incubated with diverse agents (2M Compound E, 2nM Bortezomib and 1nM Romidepsin) for 48 hours. Apoptosis detection was performed by Annexin V/PI staining and analyzed by flow cytometry. Annexin V+/PI− (lower right quadrant) areas stand for early apoptotic cells, and Annexin V+/PI+ (upper right quadrant) areas stand for late apoptotic or necrotic cells. (B) Bar graphs represent the percentages of early apoptotic cells from normal donors (Annexin V+/PI−) after treatment with therapeutic agents. Results are expressed as Mean ± SD. N=6. Supplementary Figure 2 Ex vivo triple combination treatment did not induce significant apoptosis of PBMCs from ICN-1low ATL patients. (A) PBMCs from ICN1low ATL patients were incubated with diverse therapeutic agents (2M Compound E, 2nM Bortezomib and 1nM Romidepsin) for 48 hours. Apoptosis detection was performed by Annexin V/PI staining and analyzed by flow cytometry. Annexin V+/PI− (lower right quadrant) areas stand for early apoptotic cells, and Annexin V+/PI+ (upper right quadrant) areas stand for late apoptotic or necrotic cells. (B) Bar graphs represent the percentages of early apoptotic cells from normal donors (Annexin V+/PI−) after treatment with agents. Results are expressed as Mean ± SD. N=5. 2 Supplementary Information Materials and Methods Proliferation assay ATL cell lines MT-1, ED40515(+) (abbreviated as ED+) , ED40515(-)(abbreviated as ED-), 43Tb-, LM-Y1, ED41214-, ST-1 as well as the Hut-102 (HTLV-1 infected cell line from a patient with ATL), Jurkat and HPB-ALL (T-ALL lines), Kit225 (chronic T-celllymphocytic-leukemia) were maintained in RPMI-1640 containing 10% fetal bovine serum (FBS), 100U/mL of penicillin, and 100μg/mL of streptomycin in an atmosphere containing 5% CO2. The BJ fibroblast cell line was maintained in DMEM medium following the recommendation of the ATCC (American Type Culture Collection, Manassas, Virginia, USA). ATL cells lines were obtained from Dr. Michiyuki Maeda and were previously described. The other cell lines were obtained from ATCC. Aliquots of 1×104 cells were seeded in 96-well culture plates and incubated with vehicle or Compound E, RO4929097, or Bortezomib. For Romidepsin, cells were only cultured with these agents for 6 hours, then cells were washed and cultures continued with media alone for 42 hours. The cells were pulsed after 48-hours of culture for 6-hours with 1μCi of [3H]thymidine (GE Healthcare, UK). Then, the cells were harvested with a 96-well harvester (Tomtec, Hamden, CT, USA) and counted in a β-counter (PerkinElmer Wallac, Finland). Patient materials 3 ATL patient peripheral blood samples were obtained from patients under the care of the Clinical Trials Team, Lymphoid Malignancies Branch, National Cancer Institute (NCI). This study protocol was approved by the Institutional Review Board of the NCI. Informed consent was obtained in accordance with the Declaration of Helsinki. All the ATL patients had circulating antibodies against HTLV-1, initially when recruited. Furthermore, pathological examination and FACS analysis of materials from these patients confirmed the diagnosis of ATL. RT-PCR, sequencing and tranfection Total RNA from the MT-1 cell line was reverse transcribed using a SuperScript® III First-Strand Synthesis System (Invitrogen, NY, USA). Oligonucleotide primers for PCR amplification of exon 34 of the Notch-1 gene are listed in supplementary Table-1. The amplified PCR product was purified using a QuickStepTM2 PCR-Purification-Kit (EdgeBio, Gaithersburg, MD, USA) and was directly sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, NY, USA). Human Notch-1 intracellular domain (hICN-1) expression construct EF.hICN1.CMV.GFP was obtained from Addgene(Cambridge MA, USA). Oligonucleotide primers used for PCR amplification of the cleaved portion of the Notch-1 gene are: hICN-1 Forward: 5’-GCC ACC ATG CGG CGG CAG CAT GGC CAG CTC-3’ and hICN-1 Reverse: 5’-CTT GAA GGC CTC CGG AT GCG GGC-3’. A point mutation was introduced at nt 7536 (C->T) of the Notch-1 gene using QuikChange II Site-Directed Mutagenesis Kit (Aligent Technologies, Santa Clara, CA, USA). Oligonucleotide primers use for the site directed mutagenesis of Notch-1 are: hICN-1_nt7536 (C->T), Forward: 5'- 4 ACC CCT TCC TCA CCC T GTC CCC TGA GTC CCC-3' and hICN-1_nt7536 (C->T), Reverse: 5'-GGG GAC TCA GGG GAC A GGG TGA GGA AGG GGT-3'. The mutant hICN-1 clones were sequenced directly using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, NY,USA). The amplified PCR product was subcloned into the pEF6/V5-His vector and was sequenced dirtectly using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, NY,USA). The pEF6/V5-His vectors containing either the wild type hICN-1 or the mutant hICN-1 were digested with EcoRV and BamHI and subcloned into pcDNA3.1Myc (His B) vector, then transfected into 293T cells using the Lipofectamine 200 reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacture’s instructions. Western blot analysis Samples from whole-cell lysates were prepared and proteins (30 μg) were subjected to Western blot analysis. The blots were probed with a human monoclonal antibody directed to cleaved Notch-1(Val 1744), -actin (Cell Signaling, Danvers, MA, USA). The protein bands recognized by the antibodies were visualized with an enhanced chemiluminescence Western blotting detection system (GE Healthcare, WI, USA). Therapeutic study Therapeutic experiments were performed on selected mice with palpable tumors, which occur 10 to 12 days after tumor inoculation. For the therapeutic study, Compound E was dissolved in polyethylene glycol-300 (PEG-300,VWR Scientific Products) at 10mol/Kg, and continuously administered via a subcutaneous mini-osmotic pump (ALZET, CA, 5 USA). Mini-osmotic pumps were implanted on the day treatment was initiated. The miniosmotic pumps delivered vehicle or therapeutic levels of drug at a flow rate of 0.11μL/h for 28 days. Bortezomib was provided at 0.5mg/kg/injection by intraperitoneal injection twice per week for 4 weeks. Romidepsin was administrated at 0.5mg/kg by i.p injection every other day for 4 weeks. The therapeutic groups were set up with one agent, or two or three agent combinations. One group of mice that received 200 μL PBS weekly for 4 weeks served as a control. There were 10 mice per group. The groups were randomly assigned and had comparable average levels of tumor size at the beginning of the therapeutic trials. Ex vivo cultures of PBMCs from ATL patients PBMCs from ATL patients were cultured ex vivo in RPMI-1640 with 10% FBS in the presence of inhibitors. On day 6 of the culture, 3H-TdR was added for the last 6 hours. Cell proliferation was measured by thymidine incorporation. 6