3. GPC basics
advertisement

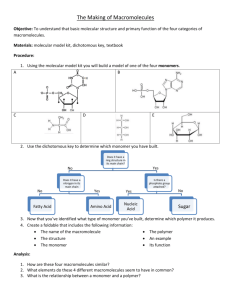
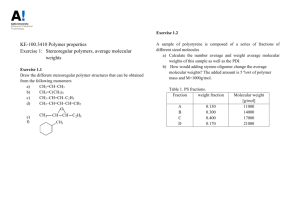
Objectives and Motivation To understand the physical principles of Size Exclusion Chromatography (SEC) and its use in the determination of the molecular weight distribution of a polymer sample. Introduction Gel Permeation Chromatography (GPC), often also known as Size Exclusion Chromatography, is a separation method for high polymers, similar to but advanced in practice over gel filtration as carried out by biochemists, that has become a prominent and widely used method for estimating molecular-weight distributions since its discovery in 1961. The separation takes place in a chromatographic column filled with beads of a rigid porous “gel”; for polymer molecular-weight determinations, highly crosslinked porous polystyrene and porous glass are preferred column-packing materials. The pores in these gels are of the same size as the dimensions of polymer materials. A sample of a dilute polymer solution is introduced into a solvent stream flowing through the column. As the dissolved polymer molecules flow past the porous beads, the can diffuse into the internal pore structure of the gel to an extent depending on their size and the pore size distribution of the gel. Larger molecules can enter only a small fraction of the internal portion of the gel, or are completely excluded; smaller polymer molecules penetrate a larger fraction of the interior of the gel. The different molecular species are eluted from the column in order of their molecular size. Therefore, the larger molecules leave the column first, and the smaller molecules, latter. A specific column or set of columns (with gels of different pore sizes) is calibrated empirically to give such a relationship, by means of which a plot of amount of solute versus retention volume, such as the ones obtained for this laboratory, can be converted into a molecular-size-distribution curve. For convenience, commercially available narrow-distribution polystyrenes are often used as standards. In order to obtain relevant information from the GPC chromatogram, it is necessary to convert it into a Molar Mass Distribution Curve (MMD) curve. Knowledge of the relationship between molar mass (M) and elution volume (Ve) – volume of solvent required to elute a particular polymer species from the point of injection to the detector – is needed to do so. Such relationship comes from the expression for total elution volume (Ve), Page 1 of 3 Ve = V0 + Vi exp(-As * L/2) (1) Where Ve = Total Elution Volume V0 = Void Volume (Volume in the system outside the porous beads) Vi = Internal Volume (Volume of solvent inside the beads) As = Surface area per unit pore volume L = Mean molecular projection of the molecule when free in solution Since L is proportional to the molar mass (M), this is the elution volume to molar mass relationship needed. This expression arises from the thermodynamics on which the Gel Permeation Chromatography is based on. First, it is known that for separations exclusively performed by size exclusion, the enthalpy change should be zero. Recalling that G = H - TS, if H=0, then G = - TS. It is also known that the relationship between the Gibbs Free Energy and the equilibrium constant is G = - RT ln K.Then if we equate this two Gibbs Free Energy expressions we have - TS = - RT ln K. Solving for K we have the following expression for K as a function of Entropy (S), K = exp (S/R). But what is all this effort in calculating K? The answer comes from the original expression for the total elution volume, which is the sum of the void volume (volume in the system outside the porous beads) and the internal volume (volume of solvent inside the beads). However, the term for internal volume is dependent of how deep the molecule penetrates inside the pores in the bed. Thus, the term K represents the fraction of internal pore volume which has been occupied by the molecule, but it is also viewed as an equilibrium contant. Then Ve = V0 + Vi*K. If the expression for K in terms of entropy is substituted in this equation, we have Ve = V0 + Vi* exp (S/R). (2) Methods of statistical thermodynamics based on models of coils entering pores of different shapes are used to obtain an alternative expression for S. The result has the general form S = -R As * L/2, which is inserted in equation (2) to give equation (1) described above. Now, GPC is not a primary method as usually practiced. As it is mentioned above, it requires calibration in order to convert raw experimental data into molecular weight distribution. The raw data in gel permeation chromatography consist of a trace of detector response (e.g. differential refractometer, continuous viscometer, etc) – which is proportional to the amount of polymer in solution – and the corresponding elution volumes. A series of commercially available anionically polymerized polystyrenes is particulary well suited to be used as standard for calibration, since anionic-polymerized polystyrene can be produced as closely monodisperse polymers and for a wide range of molecular weights. When such sample is injected into the GPC column set, the resulting chromatogram is narrower than that of a whole polymer, but is not a simple spike because of band broadening effects and dispersion along the columns, and because the polymer standard is itself not exactly monodisperse. Since the distribution is very narrow, no serious error is committed by assigning the elution volume corresponding to the peak of the chromatogram to the molecular weight of the particular polystyrene. The peaks appear in decreasing order of molecular weight, being the larger molecules the first to be eluted. A “cocktail” of several polystyrenes covering different molecular weights is usually used and result in a peak of each one of them. Once every molecular weight is assigned to its elution volume, a plot of log(M) versus elution volume can be constructed. A straight line should result and this is known as the calibration curve. After having the calibration curve ready, an analysis of the unknown polymer can be performed. Again a plot of the reading signal at the end of the set of columns versus the elution time is the result of the run. Elution time can be converted into elution volume if the pump flow is known. Note that the calibration curve to be used must cover the same range of elution volume as the known sample. It is also important to consider a baseline to which the detector response will be referred; this will be considered the origin (y=0) and the deviation will be proportional to the concentration of polymer. These recorder divisions are in fact the product NiMi, the product of the number of molecules of “i” molecular weight times “i” molecular Page 2 of 3 weight. If the unknown sample and the standard are compared at the same elution volume, it is possible to obtain Mi for such volume, applying the linear fitting expression for log(M) as a function of Ve. Once all Mi’s are determined, two columns – Ni and NiMi2 - can be obtained for each molecular weight. Therefore, Mn = Sum (NiMi) / Sum (Ni) (3) Mw = Sum (NiMi2) / Sum (NiMi) (4) PDI = Mw/Mn (5) and Where Mn stands for Number Average Molecular Weight, Mw for Weight Average Molecular Weight and PDI is the Polydispersity Index. Conclusions GPC is an extensively used technique to determine the molecular weight distribution of polymers. However, caution should be used during its application due to possible sources of error. Some of them are due to the PS standards which may not be exactly monodisperse. Other possible source of error comes from the election of the baseline; different baselines may result in different values of Molecular Weight Distribution (MWD). Errors may also arise from the precision of the detector and from the presence of branched polymers, since they are more compact than linear molecules of the same molecular weight and therefore have smaller hydrodynamic volumes. Bibliography Rudin, A. “The Elements of Polymer Science and Engineering”, Academic Press (1998), USA Billmeyer, B. “Textbook of Polymer Science” , Wiley-Interscience (1984), USA Young R and Lovell P, “Introduction to Polymers”, Stanley Thornes (2000) UK Collins, E et al. “Experiments in Polymer Science” Page 3 of 3