Clustering, Distance Methods & Ordination: Lecture Notes
advertisement
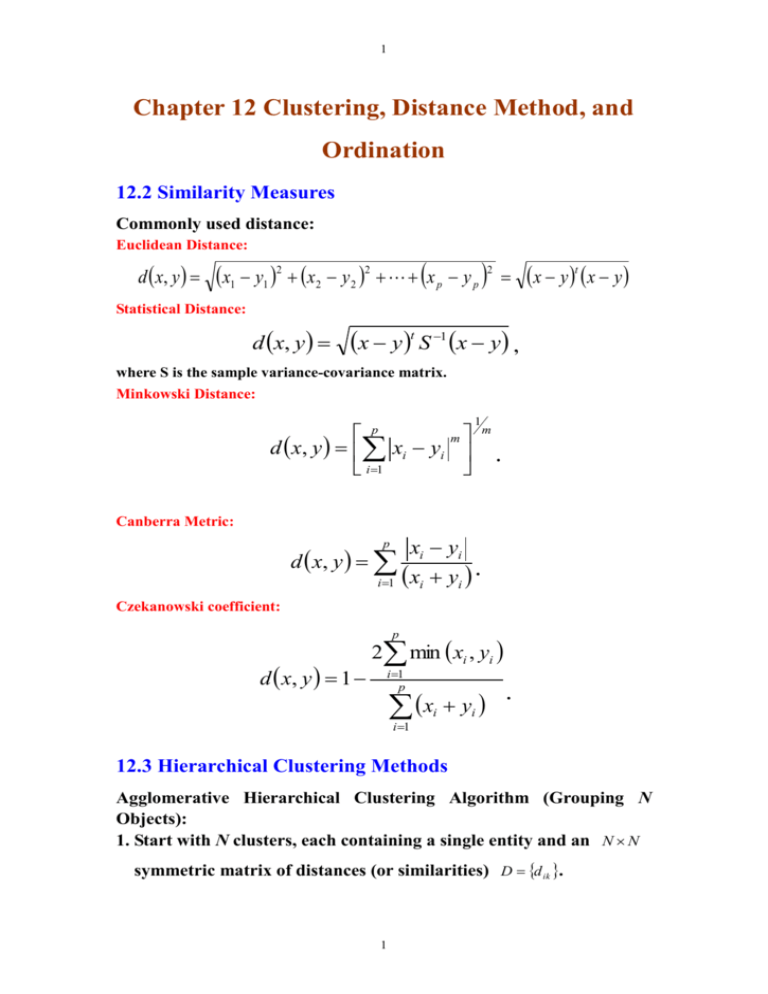
1 Chapter 12 Clustering, Distance Method, and Ordination 12.2 Similarity Measures Commonly used distance: Euclidean Distance: d x, y x1 y1 2 x2 y 2 2 x p y p 2 x y t x y Statistical Distance: d x, y x y t S 1 x y , where S is the sample variance-covariance matrix. Minkowski Distance: p m d x, y xi yi i1 1 m . Canberra Metric: p d x, y i 1 xi yi xi yi . Czekanowski coefficient: p d x, y 1 2 min xi , yi i 1 p x y i 1 i . i 12.3 Hierarchical Clustering Methods Agglomerative Hierarchical Clustering Algorithm (Grouping N Objects): 1. Start with N clusters, each containing a single entity and an N N symmetric matrix of distances (or similarities) D d ik . 1 2 2. Search the distance matrix for the nearest (most similar) pair of clusters. Let the distance between “most similar” clusters U and V be dUV . 3. Merge clusters U and V. Label the newly formed cluster (UV). Update the entries in the distance matrix (a) deleting the rows and columns corresponding to clusters U and V and (b) adding a row and column giving the distances between cluster (UV) and the remaining clusters. 4. Repeat Steps 2 and 3 a total of N-1 times. (All objects will be in a single cluster after the algorithm terminates.) Record the identity of clusters that are merged and the levels (distances) at which the merges take place. There are 3 linkage methods. The main differences among these methods are the distances between (UV) and any other cluster W. (I) Single Linkage: d UV W min d UW , d VW . (II) Complete Linkage: d UV W max dUW , dVW . (III) Average Linkage: d UV W where d ik d i ik k N UV NW , is the distance between object i in the cluster (UV) and object k in the cluster W, and N UV and N W are the number of items in clusters (UV) and W, respectively. 2











