Use of unannotated genome sequence for protein
advertisement
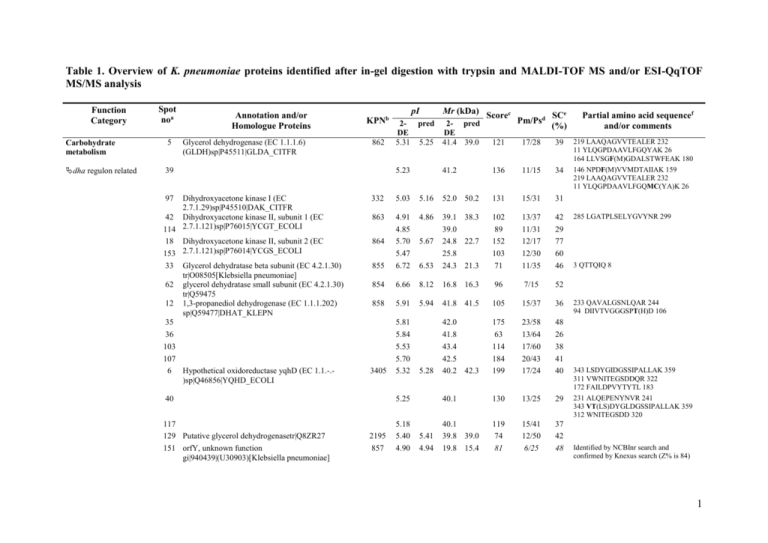
Table 1. Overview of K. pneumoniae proteins identified after in-gel digestion with trypsin and MALDI-TOF MS and/or ESI-QqTOF MS/MS analysis Function Category Spot noa Carbohydrate metabolism 5 dha regulon related 39 Annotation and/or Homologue Proteins Glycerol dehydrogenase (EC 1.1.1.6) (GLDH)sp|P45511|GLDA_CITFR 97 Dihydroxyacetone kinase I (EC 2.7.1.29)sp|P45510|DAK_CITFR 42 Dihydroxyacetone kinase II, subunit 1 (EC 114 2.7.1.121)sp|P76015|YCGT_ECOLI 18 Dihydroxyacetone kinase II, subunit 2 (EC 153 2.7.1.121)sp|P76014|YCGS_ECOLI 33 Glycerol dehydratase beta subunit (EC 4.2.1.30) tr|O08505[Klebsiella pneumoniae] 62 glycerol dehydratase small subunit (EC 4.2.1.30) tr|Q59475 12 1,3-propanediol dehydrogenase (EC 1.1.1.202) sp|Q59477|DHAT_KLEPN 35 36 103 107 6 Hypothetical oxidoreductase yqhD (EC 1.1.-.)sp|Q46856|YQHD_ECOLI pI KPNb 862 Pm/Psd SCe (%) 121 17/28 39 219 LAAQAGVVTEALER 232 11 YLQGPDAAVLFGQYAK 26 164 LLVSGF(M)GDALSTWFEAK 180 41.2 136 11/15 34 146 NPDF(M)VVMDTAIIAK 159 219 LAAQAGVVTEALER 232 11 YLQGPDAAVLFGQMC(YA)K 26 5.23 332 5.03 5.16 52.0 50.2 131 15/31 31 863 4.86 855 4.91 4.85 5.70 5.47 6.72 6.53 39.1 38.3 39.0 24.8 22.7 25.8 24.3 21.3 102 89 152 103 71 13/37 11/31 12/17 12/30 11/35 42 29 77 60 46 854 6.66 8.12 16.8 16.3 96 7/15 52 858 5.91 5.94 41.8 41.5 105 15/37 36 3405 5.81 5.84 5.53 5.70 5.32 5.28 42.0 41.8 43.4 42.5 40.2 42.3 175 63 114 184 199 23/58 13/64 17/60 20/43 17/24 48 26 38 41 40 40.1 130 13/25 29 40.1 39.8 39.0 19.8 15.4 119 74 81 15/41 12/50 6/25 37 42 48 864 40 117 129 Putative glycerol dehydrogenasetr|Q8ZR27 151 orfY, unknown function gi|940439|(U30903)[Klebsiella pneumoniae] Mr (kDa) Scorec 2- pred 2- pred DE DE 5.31 5.25 41.4 39.0 5.67 5.25 2195 857 5.18 5.40 4.90 5.41 4.94 Partial amino acid sequencef and/or comments 285 LGATPLSELYGVYNR 299 3 QTTQIQ 8 233 QAVALGSNLQAR 244 94 DIIVTVGGGSPT(H)D 106 343 LSDYGIDGSSIPALLAK 359 311 VWNITEGSDDQR 322 172 FAILDPVYTYTL 183 231 ALQEPENYNVR 241 343 VT(LS)DYGLDGSSIPALLAK 359 312 WNITEGSDD 320 Identified by NCBInr search and confirmed by Knexus search (Z% is 84) 1 Glycolysis/ gluconeogenesis 87 Phosphoglucomutase (EC 5.4.2.2) (Glucose phosphomutase) (PGM)sp|P36938|PGMU_ECOLI 15 Fructose-bisphosphate aldolase class II (EC 4.1.2.13) sp|P11604|ALF_ECOLI 17 Triosephosphate isomerase (EC 5.3.1.1) (TIM) 140 sp|Q9Z6B9|TPIS_ENTCL 19 Glyceraldehyde 3-phosphate dehydrogenase (EC 1.2.1.12)P24164_G3P_KLEPN 30 31 135 4 Phosphoglycerate kinase (EC 2.7.2.3) sp|Q8XG18|PGK_SALTY 41 112 29 Phosphoglycerate mutase 1 (EC 5.4.2.1) sp|P31217|PMG1_ECOLI 91 Phosphoglyceromutase (BPG-independent PGAM) (EC 5.4.2.1) sp|P37689|PMGI_ECOLI 2 Enolase (EC 4.2.1.11) sp|P08324|ENO-ECOLI 3 51 98 85 Pyruvate kinase I (EC 2.7.1.40) sp|P77983|KPY1_SALTY 818 5.30 5.37 57.2 58.6 72 12/39 22 482 LTAAPGD(N)GAAIGGLK 496 463 LSPEF(M)VSADTLAGDPITAR 481 253 LNLTIVNDQVDQTFR 267 2258 5.69 5.67 39.0 39.1 82 12/34 49 309 AYLQGQLGNPK 319 58 IVFSD(N)GGAAFIAGK 72 5084 5.78 1223 5.70 5.54 6.57 6.67 31.5 26.9 32.1 35.5 35.5 114 55 125 20/66 8/42 16/31 59 23 44 2259 6.25 6.07 5.91 5.14 5.02 35.6 35.9 36.2 41.6 40.5 55 73 77 216 14/80 13/38 15/64 21/29 45 36 46 66 5.07 41.6 159 23/43 66 124 YAALCDV(EMN)FVMDAFGTAHR 141 g75 4.99 5.62 5.86 41.6 32.3 28.4 112 262 13/26 25/62 32 82 161 VVPYWNETILPR 172 5033 5.33 5.66 54.0 51.7 102 15/44 37 g1949 5.28 5.33 46.5 45.5 115 13/33 56 g4554 5.22 5.17 5.09 5.61 5.66 46.1 45.9 46.0 58.4 50.7 65 58 56 53 13/32 12/46 7/22 7/32 48 32 26 24 58.7 282 4/24 19 64.9 72.7 65.4 36.0 35.4 36.4 36.1 57 178 107 86 69 9/23 6/38 14/49 12/48 10/44 15 10 48 38 29 84 5.52 308 LVSWYDNETGYSNK 321 297 AGIALNDNFVK 307 201 AQNIIPSSTGAAK 213 229 YEADLVDEAK 238 254 VATEFSETATATLK 267 204 DQLIVGGGIANTFVA 218 283 EDATIADLAVGTAAGQIK 300 319 IQ(E)EALGEQAPFD(N)GR 332 248 GVEIPVEEV(G)I 257 303 LDGTDAVF(M)LSGESAK 317 304 VANAL(V)LDGTDAVF(M) LSGESAK(AETA) 322 248 GVEIPVEEV(G)I 257 390 YFPDATILALTT 401 Pentose pathway 81 82 123 121 122 Transketolase 1 (EC 2.2.1.1) (TK 1) sp|P27302|TKT1_ECOLI 2261 Transaldolase B (EC 2.2.1.2) sp|P30148|TALB_ECOLI 760 5.48 5.52 4.95 5.07 5.04 5.67 4.99 533 TAEQLANVAR 542 321 AFPQEAAEFTR 331 2 EntnerDoudoroff pathway 149 KHG/KDPG aldolase sp|P10177|ALKH_ECOLI 421 5.41 5.50 28.5 22.4 87 11/39 58 TCA cycle 80 4087 5.66 5.83 65.6 66.0 114 17/43 30 2729 5.49 5.54 36.7 32.6 121 19/74 53 4159 5.37 5.02 70.2 70.2 67 13/46 19 2114 5.09 5.15 1282 4.87 5.13 5.06 4.77 4.70 17.9 14.9 44.9 42.0 44.1 17.0 12.0 63 78 86 58 5/33 12/40 13/35 5/21 69 28 26 52 4317 5.25 5.24 31.4 27.2 104 12/27 45 h2981 5.25 +2978 +2979 +2980 653 5.72 5.29 42.6 42.8 69 8/38 31 5.73 52.5 55.2 132 26 55 4.89 54.5 54.0 48.0 50.2 208 150 208 20/25 18/34 24/29 49 31 48 Acetate formation One-Carbon metabolism Hydrogen transport system Others Energy Metabolism ATP synthesis Fumarate reductase flavoprotein subunit (EC 1.3.99.1) sp|P00363|FRDA_ECOLI 133 Malate dehydrogenase (EC 1.1.1.37) sp|Q60150|MDH_ECOLI 79 Phosphate acetyltransferase (EC 2.3.1.8) sp|P39184|PTA_ECOLI 168 Protein yfiD sp|P33633|YFID_ECOLI 72 S-adenosylmethionine synthetase (EC 2.5.1.6) 99 sp|P04384|METK_ECOLI 163 Thioredoxin 1 sp|P00274|THIO_ECOLI 146 2-deoxy-D-gluconate 3-dehydrogenase (EC 1.1.1.125) sp|P37769|KDUD_ECOLI 113 Mannose-6-phosphate isomerase (EC 5.3.1.8) sp|P00946|MANA_ECOLI 10 48 93 43 ATP synthase alpha chain (EC 3.6.3.14) sp|P00822|ATPA_ECOLI ATP synthase beta chain (EC 3.6.3.14) sp|P00824|ATPB_ECOLI g3124 655 5.58 5.48 4.92 16 IAQFNVVSEAHNEGTIVSVS 35 402 ELAAFSQFASDLD 414 1 MQLNSTEQ(I)SN(E)LIK 13 346 DPLVVGQEHYDTAR 359 248 YTLAGTEVSALLGR 261 115 PSYEELSSSQELLETGIK 132 346 DPLVVGQEHYD 356 168 AIEHGVSYS(SGYSV)FAGVGER 183 248 YTLAGTEVSALL 259 69 Phosphate metabolism Sulfur metabolism Amino Acids Metabolism Aspartate family Lysine, ATP synthase gamma chain (EC 3.6.3.14) sp|P00837|ATPG_ECOLI 58 Inorganic pyrophosphatase (EC 3.6.1.1) sp|Q8XGI0|IPYR_SALTY 144 3-mercaptopyruvate sulfurtransferase (EC 2.8.1.2) sp|P31142|THTM_ECOLI 105 Aspartate-semialdehyde dehydrogenase (EC 1.2.1.11) sp|P00353|DHAS_ECOLI 37 2,3,4,5-trtrahydropyridine-2-carboxylate N- 654 8.55 9.11 33.1 31.6 99 12/32 38 2803 4.86 4.90 24.3 19.8 50 5/18 40 4761 4.57 4.63 34.2 31.3 61 11/56 38 3774 5.52 5.64 41.2 40.1 72 14/38 34 919 5.40 5.41 36.1 30.0 231 23/43 62 3 diaminopimelate Threonine, Glycine, Serine Cysteine 125 succinyltransferase sp|P03948|DAPD_ECOLI 9 Threonine synthase (EC 4.2.3.1) sp|P00934|THRC_ECOLI 109 2-amino-3-ketobutyrate coenzyme A ligase (EC 2.3.1.29) sp|P07912|KBL_ECOLI 131 Phosphoserine aminotransferase (EC 2.6.1.52)(PSAT) sp|P55900|SERC_SALTY 55 Serine hydroxymethyltransferase (EC 2.1.2.1) sp|P06192|GLYA_SALTY 142 Phosphoadenosine phosphosulfate reductase (EC 1.8.4.8) sp|P17854|CYSH_ECOLI 16 Cysteine synthase A (EC 4.2.99.8) sp|P11096|CYSK_ECOLI 14 166 Valine, isoleucine Leucine Arginine Histidine 83 Dihydroxy-acid dehydratase (EC 4.2.1.9) (DAD) sp|P40810|ILVD_SALTY 86 Acetolactate synthase isozyme I large subunit (EC 4.1.3.18) sp|P08142|ILVB_ECOLI 95 3-isopropylmalate dehydratase large subunit (EC 4.2.1.33) sp|P30127|LEU2_ECOLI 106 Carbamoyl-phosphate synthase small chain (EC 6.3.5.5) sp|P14845|CARA_SALTY 8 Argininosuccinate synthase (EC 6.3.4.5) sp|P22767|ASSY_ECOLI 101 134 ATP phosphoribosyltransferase (EC 2.4.2.17) sp|P00499|HIS1_SALTY 549 5.28 5.48 5.47 36.1 43.8 46.7 203 61 19/31 11/39 49 33 5039 5.74 5.65 40.8 43.5 144 19/48 38 3552 5.46 5.44 38.9 39.9 92 17/63 50 4788 5.99 6.47 43.3 45.5 107 10/16 40 1956 5.76 5.81 31.6 27.8 188 18/36 47 h3706 5.79 +3707 +3708 +3709 5.84 37.1 34.4 56 32 6/29 3/29 47 57 32 3/29 65 173 19/29 55 63 20 36 6/21 2/21 3/21 51 25 65 147 15/21 47 56 28 5/28 3/28 51 28 h3706 5.60 +3707 +3709 37.8 h3706 5.46 +3707 38.1 483 5.49 5.53 61.9 66.3 132 142 13/28 17/31 41 23 2156 5.24 5.18 56.5 59.6 89 14/35 24 4862 6.16 6.18 47.9 50.2 132 14/27 34 4818 5.63 5.08 41.9 21.3 93 8/19 35 2886 5.44 5.43 46.0 49.9 78 9/15 27 4013 5.39 5.68 5.76 46.0 36.4 33.5 78 110 17/68 14/41 40 46 271 AATNANDTVPR 281 15 ALGANLVLTEGAK 27 18 IQGIGAGFIP 27 1 MPETMSIER 9 Confirmed by NCBInr search and by Knexus search (Z% is 99) 18 IQGIGAGFIPFH(GNL)DLK 33 15 ALGANLVLTE 24 Confirmed by NCBInr search and by Knexus search (Z% is 99) Confirmed by NCBInr search and by Knexus search (Z% is 99) 246 YYEQDDASALPR 257 454 VYESQDDAVEAILGGK 469 337 FDSQALLMR 349 44 H(N)LGQPDEDDYDAIPR 58 429 SASSATGLPQVENLENK 445 4 Phenylalamine, Tyrosine and Tryptophan Tryptophan Nucleotide and Deoxyribo-nucleotide Metabolism Coenzyme and Vitamin NAD biosynthesis Vitamin C Lipid Metabolism Fatty acid biosynthesis Liposaccharide biosynthesis Genetic Information Processing DNA RNA transcription Transcription regulation Posttranscriptional processing Protein Translation 167 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (EC 4.1.2.15) sp|p00888|AROF_ECOLI h876 +875 5.74 5.42 39.9 38.8 145 Tryptophan synthase alpha chain sp|P00930|TRPA_KLEAE 111 Tryptophan synthase beta chain (EC 4.2.1.20) sp|P00932|TRPB_ECOLI 100 Phosphopentomutase (EC 5.4.2.7)(Phosphodeoxyribomutase) sp|P07651|DEOB_ECOLI 127 Ribose-phosphate pyrophosphokinase (EC 2.7.6.1) (RPPK) sp|P15849|KPRS_SALTY 143 Uridine phosphorylase (EC 2.4.2.3) (UDRPase) sp|O33808|UDP_SALTY 128 NH(3)-dependent NAD(+) synthetase (EC 6.3.5.1) (Nitrogen-regulatory protein) sp|P18843|NADE_ECOLI 21 2,5-diketo-D-gluconic acid reductase A (EC 1.1.1.-) sp|Q46857|DKGA_ECOLI 96 Biotin carboxylase (EC 6.3.4.14) (A subunit of acetyl-CoA) sp|P24182|ACCC_ECOLI 136 Enoyl-[acyl-carrier-protein] reductase [NADH](EC 1.3.1.9) sp|P29132|FABI_ECOLI 139 2-dehydro-3-deoxyphosphooctonate aldolase (EC 4.1.2.16) sp|P17579|KDSA_ECOLI 54 Single-strand binding protein (SSB) sp|P02339|SSB_ECOLI 56 Chaperone protein DnaK (Heat shock protein) sp|Q8Z9R1|DNAK_SALTI 116 DNA-directed RNA polymerase alpha chain (EC 2.7.7.6) sp|P00574|RPOA_ECOLI 159 Transcription elongation factor greA sp|Q8XGQ9|GREA_ SALTY 154 Flavoprotein wrbA (Trp repressor binding protein) sp|P30849|WRBA_ECOLI 76 Polynucleotide phosphorylase (EC 2.7.7.8) (PNPase) sp|P05055|PNP_ECOLI g, h70 5.00 5.04 +372 371 6.04 791 44 7 Trigger factor (TF) sp|Q8XFC4|TIG_SALTY Elongation factor TU (EF-TU) sp|P21694|EFTU_SALTY Confirmed by NCBInr search and confirmed by Knexus search (Z% is 82) 31.2 28.7 39 23 69 105 4/15 3/15 7/15 13/42 17 11 14 46 5.98 41.8 43.2 140 21/51 47 5.20 5.07 43.8 44.6 107 12/26 26 g4399 5.38 5.23 37.0 34.1 70 11/43 44 1574 5.88 5.89 31.4 27.2 62 12/61 50 1471 5.40 5.38 37.0 30.6 126 15/48 65 3406 5.92 5.98 31.9 30.8 176 21/59 76 125 6.48 6.10 45.5 46.3 242 23/35 51 1173 5.46 5.43 35.0 28.1 61 6/16 22 2003 5.90 5.89 33.7 31.0 128 11/20 30 5156 5.47 5.45 23.9 18.7 78 13/71 61 766 4.84 4.83 61.7 69.1 124 13/18 27 2345 5.02 4.99 39.7 32.4 68 12/62 43 2897 4.68 4.76 21.3 17.8 50 6/36 52 2234 5.87 5.84 21.2 20.9 79 9/27 41 2878 5.14 5.08 70.0 76.8 109 23/68 33 h439 4.87 4.83 51.8 48.2 241 31/70 69 115 NVALEEQAVEAVLAK 129 +438 f66 5.43 5.3 42.4 43.2 133 16/49 51 60 GITINTSHVEYDTW(P)T 74 Identified by NCBInr search and confirmed by Knexus search (Z% is 99) 5 13 Elongation factor Ts (EF-Ts) sp|Q8XGS0|EFTS_SALTY 120 121 122 77 Elongation factor G (EF-G) sp|P02996|EFG_ECOLI 78 64 61 63 50S ribosomal protein L6 sp|P02390|RL6_ECOLI 50S ribosomal protein L9 sp|Q8Z163|RL9_SALTI 50S ribosomal protein L10 (L8) sp|P17352|RL10_SALTY 27 50S ribosomal protein L7/L12 (L8) sp|P18081|RL7_SALTY 161 50S ribosomal protein L19 sp|P02420|RL19_ECOLI 24 30S ribosomal protein S6 sp|P02358|RS6_ECOLI 915 g48 +128 5.13 5.13 5.07 5.04 5.23 5.15 36.2 30.4 169 15/29 50 5.24 37.4 36.4 36.1 71.2 74.5 198 153 131 47 56 122 22/48 17/48 15/44 8/61 11/61 24/61 55 55 47 41 27 33 75 38 54 10/53 7/53 12/44 35 20 17 g48 +128 5.27 223 2827 1206 8.84 6.16 8.50 9.71 6.19 9.04 24.0 18.9 17.9 15.8 18.5 17.8 102 193 167 7/11 16/34 20/67 39 85 82 1207 4.53 4.60 17.2 12.3 - - - 882 4.63 10.62 18.6 13.1 5.31 4.93 18.7 15.7 57 90 5/22 7/26 48 57 1372 5.25 5.49 5.50 18.7 37.3 36.8 65 171 6/36 19/47 34 57 3521 5.51 5.52 51.7 48.8 75 6/8 13 2353 4.95 5.03 21.2 18.9 53 6/31 24 2767 5.69 5.70 20.0 18.2 107 10/31 50 1503 5.30 5.59 50.3 53.2 129 18/41 45 2718 5.53 5.57 42.3 42.9 70 10/34 24 2636 5.32 5.25 5.53 5.23 53.6 52.6 53.4 45.3 46.5 74 12/38 32 h2830 71.5 26 ALTEAD(N)GDIELAIENMR 42 227 VSLTGQPFVMEPSK 240 323 IATDPFVGNLTFFR 335 Confirmed by NCBInr search and by Knexus search (Z% is 96) Confirmed by NCBInr search and by Knexus search (Z% is 87) 110 TV(SL)EEAGAEL(V)EVK 12 +2829 Posttranslational processing Protein degradation 162 132 Phenylalanyl-tRNA synthetase alpha chain (EC 6.1.1.20) sp|P08312|SYFA_ECOLI 94 Seryl-tRNA synthetase (EC 6.1.1.11) sp|P09156|SYS_ECOLI 157 Peptide deformylase (EC 3.5.1.88) sp|P27251|DEF_ECOLI 156 Peptidyl-prolyl cis-trans isomerase B (EC 5.2.1.8) sp|P23869|PPIB_ECOLI 46 Outer membrane protein tolC precursor sp|P02930|TOLC_ECOLI 104 Protease degQ precursor (EC 3.4.21.-) sp|P39099|DEGQ_ECOLI 90 Aminoacyl-histidine dipeptidase (EC 3.4.13.3) 89 sp|P15288|PEPD_ECOLI 102 Peptidase B (EC 3.4.11.-) (Aminopeptidase B) sp|Q9RF52|PEPB_SALTY 4765 5.57 263 YATVAVAADQ(K)ADALK 277 167 20/40 45 6 Environmental Information Processing Membrane transport 53 Periplasmic oligopeptide-binding protein precursor sp|P23843|OPPA_ECOLI 1395 5.85 6.14 55.8 61.4 195 31/65 57 71 Periplasmic dipeptide transport protein precursor sp|P23847|DPPA_ECOLI Putative biding protein yLiB precursor sp|P75797|YLIB-ECOLI Glycine betaine-binding periplasmic protein precursor sp|P14177|PROX_ECOLI Leu/Ile/Val-binding protein precursor(LIV-BP) sp|P02917|LIVJ_ECOLI ABC transporter periplasmic binding protein ytfQ precursor sp|P39325|YTFQ_ECOLI D-methionine-binding lipoprotein metQ precursor sp|P28635|METQ_ECOLI Histidine-binding periplasmic protein precursor (HBP) sp|P02910|HISJ_SALTY PTS system, glucose-specific IIA component (EIIAGLC) sp|P02908|PTGA_SALTY Autoinducer-2 production protein luxS (EC3.13.1.-) sp|Q9L4T0|LUXS_SALTY Betaine aldehyde dehydrogenase (EC 1.2.1.8) (BADH) sp|P17445|DHAB_ECOLI Stringent starvation protein A sp|P05838|SSPA_ECOLI N-ethylmaleimide reductase (EC 1.-.-.-) sp|P77258|NEMA_ECOLI 834 5.97 6.01 50.7 59.2 109 22/58 37 4661 8.20 8.74 48.5 56.4 119 15/24 24 1055 5.56 5.44 38.0 35.5 39 8/29 27 3798 5.68 5.69 41.1 37.4 66 6/11 17 3991 5.44 5.42 39.5 34.4 105 15/57 45 3459 5.60 5.97 33.7 28.8 122 12/28 36 1828 5.60 6.03 35.4 28.3 127 15/45 53 3713 4.71 4.73 21.1 18.2 106 9/22 43 1047 5.16 5.18 22.2 19.6 111 11/30 55 2152 5.23 5.09 54.6 52.7 98 15/40 26 1147 5.29 5.22 29.9 24.2 121 13/29 56 h733 6.06 5.81 39.9 39.5 71 10/38 56 44 5/38 47 70 73 108 130 137 115 158 quorum sensing 152 Stress response 88 148 34 1 28 11 47 92 59 22 32 Alkyl hydroperoxide reductase C22 protein (EC 1.6.4.-) sp|P19479|AHPC_SALTY Alkyl hydroperoxide reductase subunit F (EC 1.6.4.-) sp|P19480|AHPF_SALTY Putative thiol-alkyl hydroperoxide reductase (EC 1.6.4.-) tr|Q8XGE8 Superoxide dismutase [Fe] (EC 1.15.1.1) sp|P40726|SODF_SALTY Superoxide dismutase [Mn] (EC 1.15.1.1) sp|P00448|SODM_ECOLI +732 +731 3981 5.05 3980 4.92 5.44 5.03 23.4 20.7 145 16/36 78 5.4 23.4 55.5 56.0 109 139 12/22 20/40 49 41 2517 5.39 5.33 5.11 5.15 55.3 56.0 25.8 22.4 97 84 112 19/50 14/38 15/54 37 29 55 724 5.70 5.76 23.4 21.2 67 6/17 33 3668 6.15 6.23 27.6 23.0 141 13/32 43 241 LAD(N)GANYGFPVNTMHIVANK 260 4 ALETAEIPGIVNDFR 18 172 EGEATLAPSLDLVG 185 94 YAMIGDPTGALTR 106 507 ASLSAFDYLIR 517 293 LTPAATEGGLHQIETAS 309 7 10 kDa chaperonin (Protein Cpn10) (groES protein) sp|P05380|CH10_ECOLI 60 kDa chaperonin (Protein Cpn60) (groEL protein) (HSP60KP) sp|O66026|CH60_KLEPN 4105 5.38 5.39 18.6 10.4 140 11/27 73 48 ILENGTVQPLDVK 60 21 SAGGIVLTGSAAAK 34 4104 4.87 4.84 55.9 57.1 209 33/58 53 426 LAGLTGQNEDQN 437 145 AQVGTISANSDEEA(TV)GK 160 83 DG(A)AAPTLSMG (GDGTTTA)TVLAQAIVNEGLK 105 160 Putative molecular chaperone (Small heat shock protein)tr|Q8ZPY6 150 GrpE protein (HSP-70 cofactor) (Heat shock protein B25.3) sp|P09372|GRPE_ECOLI 26 16 kDa heat shock protein A 164 sp|P29209|IBPA_ECOLI 147 Phage shock protein A sp|P23853|PSPA_ECOLI 138 Hypothetical oxidoreductase yajO (EC 1.-.-.-) sp|P77735|YAJO_ECOLI 141 Probable oxidoreductase ydfG (EC 1.-.-.-) sp|P39831|YDFG_ECOLI 38 outer membrane protein A precursor sp|P2401|MPA_KLEPN 49 CLPB protein (heat shock protein F84.1) 50 sp|P03815|CLPB_ECOLI 60 Protein yifE sp|P27827|YIFE_ECOLI 67 Protein yajQ sp|P77482|YAJQ_ECOLI 68 Protein ydgH Precursor sp|P76177|YDGH_ECOLI 65 Histone-like protein HLP-1 precursor sp|P11457|HLPA_ECOLI 75 Putative outer membrane protein Vprtr|Q8KR94 118 Protein StbA (ParA locus 36 kDa protein) sp|P11904|STBA_ECOLI 119 Outer membrane protein A precursor 124 sp|P24017|OMPA_KLEPN 126 Hypothetical protein yfeX sp|P76536|YFEX_ECOLI 155 Hypothetical protein yqjDsp|P42617|YQJD_ECOLI 3068 5.40 5.56 19.3 17.6 110 9/23 38 183 4.69 4.69 28.4 21.7 112 11/27 45 4204 5.40 4350 3045 5.33 5.05 5.38 5.76 5.37 5.78 17.4 15.7 17.1 30.8 25.4 34.3 34.2 112 112 142 154 10/26 6/33 14/38 17/37 45 40 51 47 3073 5.73 5.71 32.3 27.2 125 18/46 65 2588 5.43 5.42 38.6 31.0 99 16/79 57 866 5.48 339 946 2993 706 5.43 5.41 5.93 5.49 8.59 8.82 5.95 5.60 9.19 9.43 72.4 72.4 17.9 21.1 35.5 20.8 13.2 18.3 34.0 16.5 78 96 145 212 116 107 12/26 19/61 10/21 20/52 11/21 8/17 24 30 77 86 37 52 704 5214 5.09 5.03 5.14 5.02 74.8 79.5 38.2 35.3 47 89 8/23 10/28 11 32 2588 5.14 5.30 5.31 5.41 5.42 38.4 31.0 38.6 36.7 33.2 20.5 11.0 97 119 115 91 10/25 12/28 13/34 6/13 36 47 65 41 20 45 Un-categorized 3723 1639 5.27 5.56 75.1 75 GYPITDDLDIYTR 87 65 ALFATGNFEDVR 76 a Refers to the proteins labelled in Figure 1 Protein access numbers in the ProtKpn database c Score is -10*Log(P), where P is the probability that the observed match is a random event. Protein scores greater than 50 are significant (p<0.05) for searching in ProtKpn database, and scores greater than 64 (italic marked) for searching in NCBInr database. If there is also a superscript number beside the score, it represents the position of this protein in the protein candidate list. Otherwise, it is the top one. d Pm and Ps: number of peptide mass values matched and searched, respectively b 8 e SC= Sequence coverage Bold marked amino acids are from sequencing results whereas amino acids in parentheses are predicted in the ProtKpn database, italic marked are sequences originated from the homologue protein given by searching in the database SWALL of the European Bioinformatics Institute. g Refers to partial ORFs at the beginning or the end of contigs of Klebsiella pneumoniae. pI and Mr for such partial proteins are predicted by using the homologous protein from public database. h Refers to the presence of gaps or sequence errors in DNA sequences leading to incorrect prediction of ORFs. pI and Mr for such partial proteins are predicted by using the homologous protein from public database. f 9