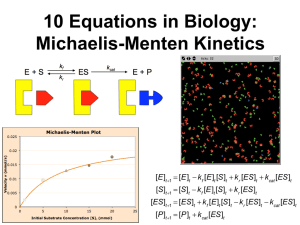
Table S1 Substrate specificity of VDH.
advertisement

Supplementary Information Functional characterization of a vanillin dehydrogenase in Corynebacterium glutamicum Wei Ding1#, Meiru Si1,2#, Weipeng Zhang1, Yaoling Zhang1, Can Chen1,2, Lei Zhang1,2, Zhiqiang Lu3, Shaolin Chen2& Xihui Shen1, 2* 1 State Key Laboratory of Crop Stress Biology for Arid Areas and College of Life Sciences, Northwest A&F University, Yangling, Shaanxi 712100, PR China; 2 Biomass Energy Center for Arid and Semi-Arid Lands, Northwest A&F University, Yangling, Shaanxi 712100, PR China; 3 College of Plant Protection, Northwest A&F University, Yangling, Shaanxi 712100, PR China. Running title: Vanillin dehydrogenase in Corynebacterium glutamicum # These authors contributed equally to this work. * Corresponding author E-mail address: xihuishen@nwsuaf.edu.cn (X.H. Shen) Key Words: Corynebacterium glutamicum; Vanillin dehydrogenase; Site-directed mutagenesis; Aromatic compound Figure S1 Multiple sequence alignment of VDHATCC13032 with identified VDHs and aldehyde dehydrogenases from other sources. The amino acid sequence of the VDH from C. glutamicum ATCC13032 was aligined to the amino acid sequence of the VDHs from Amycolatopsis sp. ATCC 39117, R. jostii RHA1, S. paucimobilis SYK6, and the amino acid sequence of the betaine aldehyde dehydrogenase from Pseudomonas aeruginosa DK2, the amino acid sequence of the aldehyde dehydrogenase from hyperthermophilic archaeon Sulfolobus tokodaii. And the site-directed residues were marked in black. Table S1 Substrate specificity of VDH. Substrates Relative activity (%) vanillin 100 3, 4-dihydroxybenzaldehyde 90.29 p-hydroxylbenzaldehyde 89.04 3-hydroxylbenzaldehyde 81.59 p-nitrobenzaldehyde 75.80 terephthalaldehyde 56.67 o-phthaldialdehyde 28.37 2,4-dichlorobenzaldehyde 55.93 cinnamaldehyde 24.40 syringaldehyde 23.31 benzaldehyde 37.93 phenylacetaldehyde 2.13 benzenepropanal 31.22 formaldehyde 4.27 aldehyde 3.11 VDH activity was determined according to the methods with substrate (1 mM), NAD+ (0.5 mM) and an appropriate amount of enzyme in potassium phosphate buffer (100 mM, pH 7.0), respectively. The activity was expressed as the percentage of the activity toward 1 mM vanillin. Table S2 Kinetics parameters of VDH towards vanillin, 3,4-dihydroxybenzaldehyde and p-hydroxylbenzaldehyde. Substrates Km(μM) kcat(s-1) kcat/Km (s-1μM-1) vanillin 26.31±3.09 21.03±1.22 0.80±0.04 3,4-dihydroxybenzaldehyde 30.44±4.11 23.42±1.78 0.77±0.04 p-hydroxylbenzaldehyde 39.11±4.89 22.92±1.80 0.59±0.02 The kinetics parameters were determined according to the standard enzyme assay procedure with 0.5 mM NAD+ and different concentrations of 3,4-dihydroxybenzaldehyde, p-hydroxylbenzaldehyde and vanillin by means of respective Lineweaver-Burk plots. The concentrations were five values ranging from 0.2 to 1 mM for substrates. Table S3 Kinetics parameters of VDH toward NAD+ and NADP+. NAD+ NADP+ Substrates Km (μM) kcat(s-1) kcat/Km (s-1μM -1) Km(μM) kcat(s-1) kcat/Km (s-1μM -1) vanillin 26.25±3.22 17.88±1.92 0.68±0.01 28.31±3.43 0.765±2.22 1.62±0.11 3,4-dihydroxy benzaldehyde 31.33±4.06 19.62±1.86 0.63±0.02 35.28±4.27 0.827±2.52 1.41±0.09 p-hydroxyl benzaldehyde 33.41±4.15 20.46±2.16 0.61±0.01 37.40±4.36 0.906±3.00 1.45±0.08 The kinetics parameters were determined according to the standard enzyme assay procedure with 1 mM 3,4-dihydroxy benzaldehyde, p-hydroxyl benzaldehyde and vanillin by means of respective Lineweaver-Burk plots. The concentrations were five values ranging from 0.125 to 1 mM for NAD+ or 0.125 to 2.5 mM for NADP+. Table S4 Relative activity of VDH and its mutants. Relative activity (%) NAD+ NADP+ 100 100 N157A 48.02 9.11 K180A 35.15 9.96 E199A 49.12 78.01 E258A 44.06 23.97 C292A 33.79 6.53 WT VDH activity was determined according to the methods with vanillin (3 mM), NAD+ (1 mM) (or NADP+ (2.5 mM)) and an appropriathe amount of enzyme in potassium phosphate buffer (100 mM , pH 7.0). The activity was expressed as the percentage of the activity toward wild type. (100%, 0. 689±0.026 U/mg). Table S5 Kinetics parameters of VDH toward NAD+ and NADP+. NAD+ Mutants Km(μM) kcat(s-1) NADP+ kcat/Km (s-1μM-1) Km(μM) kcat(s-1) kcat/Km (s-1μM-1) WT 20.36±3.65 12.96±1.32 0.64±0.04 22.36±3.01 49.80±1.86 2.23±0.19 N157A 98.79±7.53 8.94±0.96 0.09±0.01 242.29±10.11 20.14±0.90 0.08±0.01 K180A 90.33±7.36 12.54±1.08 0.14±0.01 256.37±21.36 54.66±1.38 0.21±0.01 E199A 103.29±6.01 6.30±0.61 0.06±0.01 61.23±4.29 46.92±0.78 0.77±0.04 E258A 70.19±5.25 10.26±0.66 0.15±0.01 114.33±9.48 13.20±1.26 0.12±0.01 C292A 88.95±6.35 5.76±0.42 0.06±0.01 148.40±12.32 12.72±1.20 0.09±0.01 The kinetics parameters were determined according to the standard enzyme assay procedure with 3 mM vanillin by means of respective Lineweaver-Burk plots. The concentrations were five values ranging from 0.125 to 1 mM for NAD+ or 0.125 to 2.5 mM NADP +. Table S6 Kinetics parameters of VDH toward vanillin. Mutants Km(μM) kcat(s-1) kcat/Km(s-1μM-1) WT 31.36±5.56 44.04±2.34 1.40±0.15 N157A 73.28±10.38 24.30±1.68 0.33±0.02 K180A 157.46±35.15 21.66±1.50 0.14±0.02 E199A 45.89±4.48 39.72±2.34 0.87±0.03 E258A 114.21±25.01 15.90±1.26 0.12±0.02 C292A 273.88±41.33 16.68±1.50 0.06±0.01 The kinetics parameters were determined according to the standard enzyme assay procedure with 1 mM NAD+ by means of respective Lineweaver-Burk plots. The concentrations were five values ranging from 0.2 to 3 mM for vanillin. Table S7 Bacterial strains and plasmids used in this study. Relevant characteristics Sources JM109 recAl supE44 endAI hsdR17 gyrA96 relA1 thi Δ(lac-proAB) Stratagene BL21(DE3) hsdS gal (λcIts857 ind-l Sam7 nin-5 lacUV5-T7 gene 1) Novagen RES167 Restriction-deficient mutant of ATCC 13032;Δ(cglIM-cglIR-cglIIR) This study RES167Δncgl2578 ncgl2578 deleted in RES167 This study Mobilizable vector, allows for selection of double crossover in C. Schäfer et al.1 Strains and plasmids Strains Escherichia coli Corynebacterium glutamicum Plasmids pK18mobsacB lutamicum pK18mobsacBΔncgl2578 Construct used for in-frame deletion of ncgl2578 This study pXMJ19 Shuttle vector (Ptac lacIq pBL1 oriVC. glutamicum pK18 oriVE. coli) Jakoby pXMJ19-ncgl2578 ncgl2578 cloned into pXMJ19 for complementation This study pET28a Experssion vector with N-terminal hexahistidine affinity tag Novagen pET28a-ncgl2578 pET28a derivative for expression of ncgl2578 This study Table S8 Bacterial strains, plasmids and primers used in this study. Primers Ncgl2578upFBamHI CGCGGATCCCGCCACATCACCGACGGAC Ncgl2578upR GGATCGGCATCAAGCGCAGC Ncgl2578dwF GCTGCGCTTGATGCCGATCCCTCCACCCACTGACCTCCG Ncgl2578dwRHindIII CCCAAGCTTTGCACTTCCCGGAGGCTACC Ncgl2578FBamHI CGCGGATCCATGACTGCAACATTTGCTG Ncgl2578RKpnI/SalI CCAGGTACCGTCGACTTAGCTGCGCTTGATGCCG Ncgl2578rbsFBamHI CGCGGATCCAAAGGAGGACAACCATGACTGCAACATTTGCTG N157AF TGATTAGTCCATGGGCTTTCCCACTGAACCT N157AR AGGTTCAGTGGGAAAGCCCATGGACTAATCA K180AF CAACGCCGTAGTGATTGCGCCTGCGAGTGATACCCCAGTTAC K180AR GTAACTGGGGTATCACTCGCAGGCGCAATCACTACGGCGTTG E199AF GCACGAATCTTTGAGGCGGCCGGAGTTCCTGC E199AR GCAGGAACTCCGGCCGCCTCAAAGATTCGTGC E258AF CAATGAAAACTGTTGCACTAGCGCTCGGTGGCAACGCGCCG E258AR CGGCGCGTTGCCACCGAGCGCTAGTGCAACAGTTTTCATTG C292AF ACCAGGGACAGATTGCTATGTCAATCAACCG C292AR CGGTTGATTGACATAGCAATCTGTCCCTGGT Underlined sites indicate restriction enzyme sites added for cloning. Mutated bases in the primers are shown in boldface and the ribosome binding site (rbs) is given in italic. References: 1. Schäfer, A. et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145,69-73 (1994).