SUPPLEMENTARY MATERIAL
advertisement

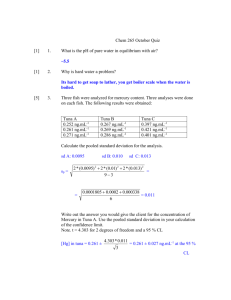
1 SUPPLEMENTARY MATERIAL 2 3 Effects of epigallocatechin gallate on the cell wall structure of 4 Mycobacterial smegmatis mc2155 5 Tieying Suna#, Biaojie Qinb#, Mingchuan Gaoa, Yuling Yina, Changyuan Wangc, Shizhu Zanga, Xinli Lia , 6 Cuili Zhanga, Yi Xina and Tao Jianga* 7 a 8 Molecular biology Department, Dalian Medical University, Dalian 116044, China;c Clinical 9 Pharmacology Department, College of Pharmacy in Dalian Medical University, Dalian 116044, China. 10 Department of Biotechnology, Dalian Medical University, Dalian 116044, China;b Biochemistry and *Corresponding Author. E-mail:jiangtaodl@163.com. 11 Abstract 12 Epigallocatechin gallate (EGCG) is the main component of green tea extracts, that 13 inhibits the growth of M.smegmatis mc2155, and the mechanism is not clear. This study 14 showed the effects of varying concentrations of EGCG on the growth of M. smegmatis 15 using ELISA analysis. The changes of EGCG content and EGCG stucture in LB 16 medium with mc2155 were identified by HPLC and LC/MS. Transmission electron 17 microscopy (TEM) provided a detail characterization of the cell envelope structure. As 18 a result, the most optional inhibition concentration was determined to be 20μg.ml-1. 19 Mass analysis found that most of EGCG was transferred into its isomeride in LB 20 medium while EGCG treatment for 18h, but inhibition effects for mc2155 had yet been 21 maintained. There were striking changes in the cell wall envelope profiles after EGCG 22 treatment for 18h. The cell wall appeared to have a less electron translucent zone, turn 23 rougher and thicker. The results show that EGCG impacts the integrity of 24 mycobacterial cell wall, and is likely be a better prophylactic agent against TB. 25 Key words: epigallocatechin gallate; M. smegmatis mc2155; antimicrobial effect; the 26 cell wall structure 27 1 Experimental 2 Antimicrobial circle experiment 3 This Drug sensitivity test adopted paper disc agar diffusion method (K-B method) to 4 observe antimicrobial functions. Ethambutol (EMB) with final concentration of 48 μg.ml-1 5 was regarded as positive control, phosphate salt buffer and ampicillin with concentration of 6 100 μg.ml-1 as negative control of antimicrobial effects. EGCG treatments were initiated by 7 adding filtered stock solution of EGCG (10 mg ml-1) to achieve the final concentration of 8 10 μg ml-1 and 20 μg ml-1 respectively. After using 0.22 μm filter, and then put paper disc 9 into every tube with different treated solution, keeping 24 h at 4℃. The plates above 10 inverted at 37℃ for 48h after placing at room temperature for 15 min, observing results. 11 Repeated the above-mentioned experiment three times, measured the antimicrobial circle 12 diameter. 13 14 The effect of EGCG on the growth of M.smegmatis mc2155/ pSMT1 M.smegmatis mc2155 strains electronfected pSMT1 was grown in LB broth containing 15 0.05% Tween 80 and 100 μg ml–1 hygromycin at 37 °C. EGCG treatments were initiated by 16 adding filtered stock solution of EGCG (10 mg ml–1) to achieve the different final 17 concentration. The plasmid DNA called pSMT1 was constructed by Prof..Douglas Young’s 18 lab, and it may produce bioluminescence. Bioluminescence has been early employed as a 19 reporter to detect the presence of low levels bacterial contamination or infection. Emission 20 of light is dependent on the presence of a cofactor, ATP or reduced flavin mononucleotide 21 (FMNH2) which is found only in living cells. Dead cells are no longer able to produce the 22 cofactor and a corresponding decline in luminescence. In this study, the growth curve of 23 M.smegmatis mc2155/pSMT1 was made according to the power of luminescence. A 24 calibration curve with M.smegmatis/ pSMT1 confirmed linearity, and 0.1 ml of 1% n-decyl 25 aldehyde in DMSO as substrates(Eklund et al. 2010). M.smegmatis mc2155/ pSMT1 was 26 seeded in per well of white opaque 96 well plates. Luminescence was measured once every 27 four hours until EGCG treatment for 24 h using SpectraMax L chemiluminescence ELISA 28 reader. 29 30 Detecting behaviors of EGCG in LB medium through HPLC and LC/MS 1 EGCG treatment were initiated to achieve final concentration of 20 μg ml-1, every 2 other 6 h to collecte supernatant through centrifuging at 5000 rpm for 10 min, and then 3 filtered with 0.22 μm filter. EGCG content in LB medium was analyzed through Binary 4 HPLC Pump (JEOL) with Hypersill BDS C18 (250 mm×4.6 mm.5μm) packed 5 chromatographic column. With 2% acetic acid as mobile phase A and with 6 chromatographic pure acetonitrile as mobile phase B, start at 5% acetonitrile and progress 7 30 %,70 % to 100 % acetonitrile over 5-20 min for gradient elution program, set detection 8 wavelength as 280 nm ,flow velocity as 0.8 ml/min and column temperature as 35℃. 9 LC/MS were acquired to identify behaviors of EGCG in LB medium. 10 Transmission electron microscopy (TEM) analysis 11 In the early exponential phase of growth (A600≈0.3), EGCG treatments were initiated to 12 achieve respectively the final concentration of 10 μg ml-1, 20 μg ml-1 and 50 μg ml-1 in LB 13 medium for 18 h. And then the cells centrifuged for 10 min and collected cells for fixing. 14 The fixed cells in 2.5% glutaraldehyde were rinsed with 0.2 M phosphate salt buffer 15 (pH=7.4), followed by dehydration with gradient alcohol. Cells were embedded with epon 16 embedding kit and cut into ultrathin sections. The sections were stained with 2% uranyl 17 acetate for 30 min and lead citrate for 20 min and finally examined with a JEM-2000EX 18 transmission electron microscope (JEOL). 19 20 1 2 3 Figure S1..Antibacterial circle experiment of green tea extracts and EGCG against 4 M.smegmatis mc2155. Effect of different material applied to a blank filter disk on an 5 agar plate inoculated with M.smegmatis mc2155; a-c represented respectively 6 phosphate salt buffer, 48 μg ml-1 EMB; 100 μg ml-1 Ampicillin; Panel A, 1 and 2 7 represented 50μg.ml-1 and 100μg.ml-1 green tea extraction; Panel B, 3 and 4 8 represented 10 μg ml-1 and 20 μg ml-1 EGCG. 9 10 11 12 13 Figure S2 The growth curve of M.smegmatis mc2155/pSMT treated by EGCG 1 Figure S3-A 2 3 4 5 Figure S3-B 1 Figure S3-C 2 3 Figure S3 HPLC and LC/MS analysis of EGCG behaviors in LB broth. 4 Figure S3-A Detecting the change of EGCG content through HPLC analysis. EGCG 5 content in LB broth was gradually decreased over time periods, and it cannot nearly 6 be detected while EGCG treatment for 30h. 7 Figure S3-B Analyzing the proportion of EGCG isomerides by HPLC. B-1, HPLC analysis 8 of EGCG standard; B-2, HPLC analysis of EGCG treated for 18h in LB broth with 9 mc2155. 10 Figure S3-C The identification of chemical structure of EGCG through LC/MS analysis. 11 C-1, mass spectrum of EGCG standard, and the peak was at m/z 457 [M-H]- 12 corresponding to EGCG with the relative molecular mass of 458.38. C-2, LC/MS 1 analysis of the peak at 3.35 min for EGCG treatment group. One peak was at m/z 453 2 [M-4H]- corresponding to EGCG with the relative molecular mass of 458.38, another 3 was m/z 382.9 which is not identified. C-3, LC/MS analysis of the peak at 4.45 min 4 for EGCG treatment group. A series of peaks at m/z 473.2, 489.4, and 498.9 might 5 correspond to ion adducts of EGCG. 6 7 8 9 Figure S4. Morphological changes of M.smegmatis mc2155 by EGCG treatment through TEM. EGCG treatments were initiated to achieve respectively the final concentration 10 of 10 μg ml-1(B), 20 μg ml-1 (C) and 50 μg ml-1 (D) in LB medium for 18 h. A, the 11 cell envelope structure of M.smegmatis mc2155; B, the cell envelope structure under 12 the treatment of 10 μg ml-1 EGCG, and some materials were dropped from the cell 13 wall; C, the cell envelope structure under the treatment of 20 μg ml-1 EGCG, and the 14 separation of the cytoplasm and cell membrane was appeared; D, the cell envelope 15 structure under the treatment of 50 μg ml-1 EGCG, and the cell wall was likely to 16 result from structural damage and turn thicker. 17 18 1 Reference 2 Eklund DA, Welin T, Schon O, Stendahl K, Huygen, M. Lerm. 2010. Validation of a 3 medium-throughput method for evaluation of intracellular growth of Mycobacterium 4 tuberculosis. Clin. Vaccine Immunol. 17:513-517. 5 6 7 8 9 10