Molecular Docking Tutorial: HIV Protease Inhibitor XK263
advertisement
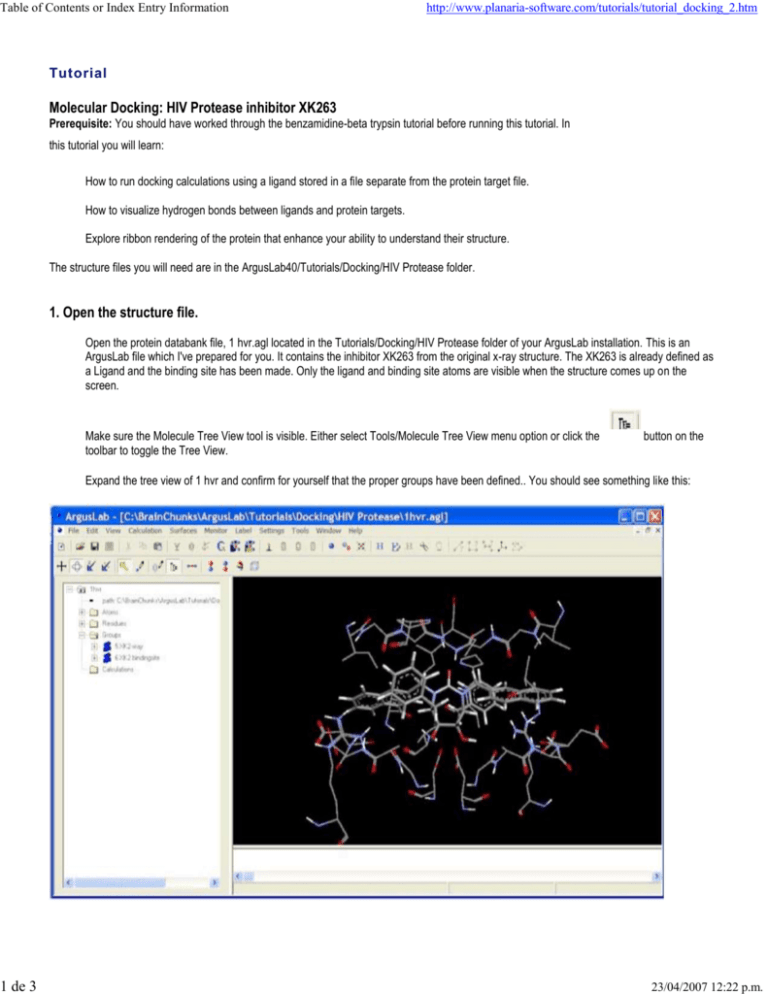
Table of Contents or Index Entry Information 1 de 3 http://www.planaria-software.com/tutorials/tutorial_docking_2.htm Tutorial Molecular Docking: HIV Protease inhibitor XK263 Prerequisite: You should have worked through the benzamidine-beta trypsin tutorial before running this tutorial. In this tutorial you will learn: How to run docking calculations using a ligand stored in a file separate from the protein target file. How to visualize hydrogen bonds between ligands and protein targets. Explore ribbon rendering of the protein that enhance your ability to understand their structure. The structure files you will need are in the ArgusLab40/Tutorials/Docking/HIV Protease folder. 1. Open the structure file. Open the protein databank file, 1 hvr.agl located in the Tutorials/Docking/HIV Protease folder of your ArgusLab installation. This is an ArgusLab file which I've prepared for you. It contains the inhibitor XK263 from the original x-ray structure. The XK263 is already defined as a Ligand and the binding site has been made. Only the ligand and binding site atoms are visible when the structure comes up on the screen. Make sure the Molecule Tree View tool is visible. Either select Tools/Molecule Tree View menu option or click the toolbar to toggle the Tree View. button on the Expand the tree view of 1 hvr and confirm for yourself that the proper groups have been defined.. You should see something like this: 23/04/2007 12:22 p.m. Table of Contents or Index Entry Information 2 de 3 http://www.planaria-software.com/tutorials/tutorial_docking_2.htm 2. Examine the hydrogen bonding in the x-ray structure. Right-click on the "XK2-xray" ligand, in the Groups folder, and select "Show hydrogen bonds". You should see several red-lines appear between the XK263 ligand and the protein. These are the hydrogen bonds between ligand and protein. Zoom the structure to bring some of the h-bonds into clearer view. Right-click on one of the red-lines and select "Hydrogen bond info...". This will bring up an info dialog with information about this particular h-bond. Notice that there are good hydrogen bonds between the two hydroxyl groups of the cyclic urea ring and the two catalytic aspartic acid residues (Asp 125 and Asp 25) and , and between the keto group of the cyclic urea and the peptide nitrogen of Ile 50. You can change the way hydrogen bonds are detected, and their color, by selecting the Settings/Monitors... menu option. Try changing the color of the h-bonds. If you change the h-bond distance value, you will then need to remove the h-bonds and re-calculate them. You can turn off the display of h-bonds by selecting the Monitor/Remove Hydrogen Bond Monitors or the Monitor/Remove All Monitors menu options. 3. Dock another copy of XK263 into the binding site. Open the file xk263.agl located in the Tutorials/Docking/HIV Protease folder. I have already created a Ligand Group for this structure and you should verify this by looking in the Groups folder under the xk263 molecule in the Tree View. Bring up the Dock Settings dialog box by selecting the Calculation/Dock a Ligand... menu option or clicking on the toolbar. button on the Under the Ligand drop-box, you should select the ligand to dock as "XK263 inhibitor" This will dock the ligand in xk263.agl into the binding site of 1 hv3.agl. Set the Binding Site Bounding Box size to 18, 18, 22 Angstroms for x,y,z respectively, by typing these dimensions into the edit boxes. Make sure ArgusDock is the docking engine, the calc type is Dock, and the Ligand is flexible options are selected. Click run. The first thing you will see is that ArgusLab copies the "XK263 inhibitor" Group from xk263.agl into 1 hvr.agl. This newly added ligand will then be docked. The final result is 1 hvr containing the additional ligand. You can dock multiple ligands from multiple sources into any molecule that has a binding site defined, thus allowing for the overlapping of multiple ligands to see common modes of binding, etc. If you left-click on the "XK2-xray" and "XK263 inhibitor" groups in the 1 hvr Group folder, then right-click on the Groups tab and select "Calc RMSD position between two similar Groups", you should see the newly docked ligand is quite close to the x-ray structure (RMSD should be < ~1.5 angstroms). Right-click on the binding site in the Groups folder and select the "Hide" option. Do the same thing for the "XK2-xray" ligand. Now only the newly docked "XK263 inhibitor" is visible. Right-click on "XK263 inhibitor" and select "Show Hydrogen Bonds" You should see just the ligand and several lines emanating from it. If you want to see just the protein residues that participate in the h-bonds, right click on any h-bond line and select the "Show All Hydrogen Bonds' Residues" option. This will make just the h-bonding residues from the target visible along with the already visible ligand. Note that these are essentially the same as the x-ray ligand's h-bonds. 4. Viewing the protein with ribbons. HIV protease is a homo-dimeric enzyme. You can visualize the overall structure of the entire protein ligand complex by rendering the protein with ribbons turned on. Select the "View/Render Protein as Cartoon Ribbon" menu option. Then select the "View/Color Ribbon By Molecule" menu option. You may have to zoom the structure out a bit to get it all on the graphics screen. You should see something like this: 23/04/2007 12:22 p.m. 2 de 3 The two subunits of this important enzyme are easily seen. Notice, also, how the two catalytic aspartic acid residues come from analogous positions on each of the two subunits. 23/04/2007 12:22 p.m.