Supplementary Information (doc 95K)
advertisement

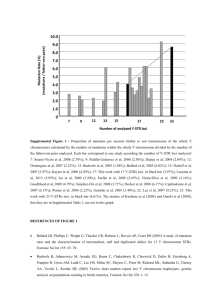
Supplemental Material Vos and Didelot ‘A comparison of homologous recombination rates in bacteria and archaea’ A. Supplemental Data for Table 1 B. Excluded Datasets C. References A. Supplemental Data for Table 1 Bacillus spp. Strains of three species (B. cereus, B. thuringiensis and B. weihenstephanensis) isolated from a forest near Versailles, France (Sorokin et al., 2006). Six loci were sequenced (clpC, dinB, gdpD, panC, purF and yhfL). Strains not isolated from the forest site were excluded. All three species live in soil. B. thuringiensis is an insect pathogen. B. cereus and B. thuringiensis can be opportunistic pathogens, B. weihenstephanensis is not associated with disease. A UK population of phylloplane B. thuringiensis was re-analyzed and found to have an r/m value of 2.0 (CI 1.2 – 3.1) (Bizarri et al., 2008). Seven loci were sequenced (glp, gmk, ilv, pta, pur, pyc and tpi) in 17 isolates. Bartonella henselae This Bartonella species is mainly associated with felines and can cause Cat Scratch Disease as well as other complications in humans. Isolates form felines and humans from a wide geographical range (Arvand et al., 2007). Seven loci sequenced (batR, gltA, ftsZ, groEL, nlpD, ribC, rpoD). Bordetella pertussis 1 Causative agent of whooping cough (pertussis) in humans. Worldwide clinical isolates, seven loci sequenced (adk, fumC, glyA, tyrB, icd, pepA and pgm) (Diavatopoulos et al., 2005). Brachyspira sp. Anaerobic intestinal spirochaetes recovered from pigs, dogs, birds and a mouse in seven European countries, USA and Canada. Species classification difficult. Seven loci used (adh, pgm, est, glp, gdh, thi, alp); no tests for recombination in original paper (Rasback et al., 2007). Campylobacter insulaenigrae Isolated from young, stranded northern elephant seals in California (Stoddard et al., 2007). Seven loci sequenced (aspA, atpA, glnA, glyA, pgi, pgm and tkt). Campylobacter jejuni British isolates from a variety of sources downloaded from pubmlst.org. Seven loci sequenced (aspA, glnA, gltA, glyA, pgm, tkt and uncA). Chlamydia trachomatis A small set of clinical isolates isolated from urogenital and eye (trachoma) infections (Pannekoek et al., 2008). Seven loci sequenced (gatA, oppA, hflX, gidA, enoA, hemN and fumC). Clostridium difficile Worldwide isolates from humans and several animal species (Lemee et al., 2004). Six loci analyzed (aroE, dutA, gmk, soda, recA and tpi). Not all alleles were available for a seventh locus ddl. Enterococcus faecalis 2 Clinical isolates, only those STs present in Spain were included; seven loci sequenced (gdh, gyd, pstS, gki, aroE, xpt and yqiL) (Ruiz-Garbajosa et al., 2006). Enterococcus faecium Dutch isolates from poultry and humans all belonging to AFLP genogroup B (Homan et al., 2002). Seven loci sequenced (adk, atpA, ddt, gyd, gdh, purK and pstS). Escherichia coli Isolates from sand and water in the wave-wash zone of six sites along Lake Huron and the St. Claire River in Michigan, USA (Walk et al., 2007). Seven loci sequenced (adk, fumC, gyrB, icd, mdh, purA and recA). Flavobacterium psychrophilum Worldwide collection of fish pathogen samples (from different host species). Seven genes sequenced (atpA, dnaK, fumC, gyrB, murG, trpB and tuf) (Nicolas et al., 2008). Haemophilus influenzae The first 50 ST’s in the www.mlst.net database (Meats et al., 2003). Effect of population structure unlikely as many ST’s occur globally. Seven loci sequenced (adk, atpG, frdB, fucK, mdh, pgi and recA). The dataset consists of different capsule types as well as some unencapsulated isolates. Non-encapsulated strains rarely cause infection whereas capsulated strains are a significant source of disease. It has been suggested that the former may recombine more frequently than the latter (Meats et al., 2003; Perez-Losada et al., 2006); this observation would be expected when disease-associated strains have epidemic population structure. Haemophilus parasuis 3 Two datasets were analyzed. One cluster of 79 mainly non-pathogenic isolates (Table 1) and a cluster of 27 mainly pathogenic isolates (Olvera et al., 2006). An r/m value of 5.5 (CI 2.6 – 32.7) was found for the latter group. No information was available on capsule production. Seven loci sequenced (atpD, infB, mdh, rpoB, 6pgd, g3pd and frdB). No congruence of individual gene trees in original publication (Olvera et al., 2006). Halorubrum sp. Isolates from different salterns at Santa Pola near Alicante, Spain. Four loci sequenced (atpB, ef-2, radA and secY). Original paper reports linkage equilibrium on the basis of the Index of Association test (Papke et al., 2004). Helicobacter pylori European isolates from pubmlst.org. Seven loci sequenced (atpA, efp, mutY, ppa, trpC, ureI and yphC). Klebsiella pneumophila Clinical (nosocomial) isolates from around Europe (Diancourt et al., 2005). Seven loci sequenced (rpoB, gapA, mdh, pgi, phoE, infB and tonB). Lactobacillus casei Isolates classified as L. casei and L.paracasei (which cluster together), mostly from (diary) food products from France (Diancourt et al., 2007). Also found in the human intestinal tract; exact natural habitat unclear. Seven loci were sequenced (fusA, ileS, lepA, leuS, pyrG, recA, recG). Legionella pneumophila Isolates from a local area around Valencia, Spain experiencing regular outbreaks (Coscolla and Gonzalez-Candelas, 2007). Fourteen sequence fragments available, but many consist of 4 very short housekeeping gene fragments and longer intergenic spacer regions. Therefore, only two gene fragments, asd and mip, were used. Low recombination rate in accordance with a study on a large collection of global isolates (Edwards et al., 2008). Leptospira interrogans Clinical (leptospirosis) and rodent isolates from Thailand (Thaipadungpanit et al., 2007). Seven loci sequenced (pntA, sucA, pfkB, tpiA, mreA, glmU and fadD). Listeria monocytogenes Spanish isolates from food and animal and human clinical sources (Salcedo et al., 2003). Seven loci sequenced (abcZ, bglA, cat, dapE, dat, ldh and ldkA). Mastigocladus laminosus Isolate collected worldwide from alkaline hotsprings (Miller et al., 2007). Four loci sequenced (argS, nifH, devH and narB). For the last three loci, only parts of the available sequences were used due to large gaps. Microcoleus chthonoplastes Isolates from two mud flat populations in the North Sea and one population in the Baltic Sea (Lodders et al., 2005). Two loci were used (KaiC and PetB/D); the available intergenic spacer sequence was not used. Microcystis aeruginosa Isolates from Japan and mainland East-Asia as well as one British and one Canadian isolate. All five Microcystis morphospecies present. Seven loci were used (ftsZ, glnA, gltX, gyrB, pgi, recA and tpi). Evidence for recombination found in original paper using the SH-test but not the Index of Association test (Tanabe et al., 2007). 5 Moraxella catarrhalis The first 50 STs from the http://web.mpiib-berlin.mpg.de/mlst database that occur in Europe. Seven loci sequenced (abcZ, adk, efp, fumC, glyBETA, mutY, ppa and trpE). The finding of high HRR contrasts with a previous study (Perez-Losada et al., 2006) that utilized coalescent methods on the whole Berlin dataset. This could potentially be due to inclusion of geographically isolated groups between which no recombination occurs. Mycoplasma hyopneumoniae Can cause enzootic pneumoniae in swine. Most strains from Switzerland; several very closely related strains from other countries included as well (four outgroup STs omitted) (Mayor et al., 2007). Seven loci sequenced (efp, metG, pgiB, recA, adk, rpoB and tpiA) Myxococcus xanthus A previous study provided evidence that populations of this soil bacterium are unstructured below a scale of 102 – 103 km (Vos and Velicer, 2008) and so all distinct ST’s collected below such a regional scale (SW Germany) were used. Five loci were sequenced (clpX, csgA, fibA, icd and sglK). Neisseria lactamica Seven loci (abcZ, adk, aroE, fumC, gdh, pdhC and pgm) were analyzed in isolates from around the world (www.pubmlst.net). Neisseria meningitides Commensal, non-disease associated STs from the Czech republic (Jolley et al., 2005). Loci as in N. lactamica. Oenococcus oeni 6 Isolates from a variety of wine industry strain collections from all over the world (de Las Rivas et al., 2004). Five loci analyzed (gyrB, ddl, pgm, recP and mleA). Pelagibacter ubique (SAR 11) Isolated from a 2 liter sample of sea water collected of the Oregon Coast. Eight loci used (HSP60, ATPDH, ACoAAT, recA, OR, DPoIIIα, RPoIβ). A variety of tests, including the coalescent-based program LDHat, provide strong evidence of very high recombination rates in the original paper (Vergin et al., 2007). Plesiomonas shigelloides Five loci sequenced (fusA, leuS, pyrG, recG and rpoB). Isolates from, sewage, lakes, rivers and a variety of animals. Only European isolates used for the analysis. Evidence for extensive recombination presented in the original publication (Salerno et al., 2007). Porphyromonas gingivalis Global collection of periodontal pathogens. Seven loci sequenced (gdpxJ, pga, hagB, mcmA, recA, ftsQ and pepO). No tests for recombination in the original publication (Enersen et al., 2006). Pseudomonas syringae A collection of isolates from the foliage of a range of plant species. Isolates from across the world, but little evidence of spatial structuring (Sarkar and Guttman, 2004). Four loci sequenced (gapA, gltB, gyrB and rpoD). Sequences of additional strains provided by D. Guttman. Pseudomonas viridiflava Most isolates from the USA, some European isolates included as well (the original publication reports little geographic clustering) (Goss et al., 2005). Isolates found on a variety 7 of host plants. Three loci sequenced (gyrB, purA and aputative radical sterile alpha motif domain protein). Ralstonia solanacearum Worldwide collection isolated from a variety of crops (Castillo and Greenberg, 2007). Seven loci sequenced (gyrB, adk, gdhA, gapA, ppsA and fliC). Rhizobium gallicum R. gallicum sensu lato Group I strains isolated from around the northern hemisphere (Silva et al., 2005). Three loci sequenced (glnII, atpD and nodB). Salmonella enterica The first 50 european isolates from the http://web.mpiib-berlin.mpg.de/mlst database. Seven loci sequenced (aroC, dnaN, hemD, hisD, purE, sucA and thrA). High recombination rate is in accordance with data on Salmonella enterica subspecies I (Octavia and Lan, 2006). Staphylococcus aureus Clinical isolates from Oxfordshire (Enright et al., 2000). Seven loci sequenced (arcC, aroE, glpF, gmk, pta, tpi and yqiL). Streptococcus pneumoniae A distinct clade (Group 1 in (Hanage et al., 2005)) of typeable and nontypeable strains from middle ear fluid and the nasopharynx. Six loci used (aroE, gdh, gki, recP, spi and xpt). Streptococcus pyogenes Group A Streptococci (GAS) are predominantly human pathogens. Seven loci (gki, gtr, murl, mutS, recP, xtp and yqiL) were sequenced in upper respiratory tract-, invasive- and impetigo isolates from the USA (the first 50 ST’s listed in the original study (Enright et al., 2001)). 8 Sulfolobus islandicus Isolates from two hot springs (approx. 25 m apart) near the Mutnovsky Volcano on the Kamchatka peninsula (Whitaker et al., 2005). Five loci used (hypothetical molydobterine oxidoreductase, hypothetical acetyl Coa acetyl-transferase, hypothetical thermosome gamma subunit, hypothetical transcription initiation factor and putative ATP-dependent helicase) (one locus excluded because of evidence for selection). Vibrio parahaemolyticus A subset of 20 environmental isolates from the United States Pacific Coast was analyzed. Seven loci (dnaE, gyrB, recA, dtdS, pntA, pyrC and tnaA) present on both chromosomes were used. The original paper (Gonzalez-Escalona et al., 2008) reports high recombination rates on the basis of a variety of tests. Vibrio vulnificus A distinct clade of mainly environmental isolates from around the world (Bisharat et al., 2007). Five loci from the larger chromosome used (glp, gyrB, mdh, metG and purM). Wolbachia b complex Five loci sequenced (gatB, coxA, hcpA, fbpA and ftsZ) in symbionts from a variety of hosts (Baldo et al., 2006). Insects can host multiple Wolbachia strains and strains are not limited to a single host species. Classification of strains into species is still unresolved. The analysis was performed on MLST complex b (with strains sharing minimal 3 identical alleles). Several studies have documented homologous recombination in this species (Baldo et al., 2006; Baldo et al., 2006; Jiggins et al., 2001). Yersinia pseudotuberculosis 9 Isolates from Asia isolated from humans, animals and vegetables. Seven loci sequenced (adk, argA, aroA, glnA, thrA, tmk and trpE); sequences from http://web.mpiib-berlin.mpg.de/mlst/. B. Excluded Datasets Several datasets were excluded because credibility intervals were extremely wide and so mean r/m was not informative: Acinetobacter baumannii: Clinical isolates from four European countries as well as several reference strains (Bartual et al., 2005). Seven loci sequenced (gltA, gyrB, gdhB, recA, cpn60, gpi and rpoD). Burkholderia amfibaria: Eleven isolates from maize rhizosphere, Italy (Dalmastri et al., 2007). Seven loci sequenced (atpB, gltB, gyrB, recA, lepA, phaC and trpB). Burkholderia multivorans: Worldwide collection of mainly pathogenic (CF lung) strains (www.pubmlst.net). Loci as in B. amfibaria. Lactobacillus plantarum: Isolates collected in different countries, in different years (as far back as 1948) from different sources (wine, foods, saliva, silage) (de Las Rivas et al., 2006). (Sample thus not representative of a natural population.) Six loci sequenced (pgm, ddl, gyrB, purK1, gdh and mutS). Pseudomonas aeruginosa: The set consists of the first 50 ST’s from the UK archived at www.pubmlst.net. Mainly from clinical samples, with several environmental isolates. Seven loci sequenced (acsA, aroE, guaA, mutL, nuoD, ppsA and trpE) (Curran et al., 2004). 10 C. References Arvand M, Feil EJ, Giladi M, Boulouis HJ, Viezens J (2007). Multi-Locus Sequence Typing of Bartonella henselae Isolates from Three Continents Reveals Hypervirulent and FelineAssociated Clones. PLoS ONE 2: e1346. Baldo L, Bordenstein S, Wernegreen JJ, Werren JH (2006). Widespread recombination throughout Wolbachia genomes. Mol Biol Evol 23: 437-49. Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR et al (2006). Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol 72: 7098-110. Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodriguez-Valera F (2005). Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol 43: 4382-90. Bisharat N, Cohen DI, Maiden MC, Crook DW, Peto T, Harding RM (2007). The evolution of genetic structure in the marine pathogen, Vibrio vulnificus. Infect Genet Evol 7: 685-93. Bizarri MF, Prabhakar A, Bishop AH (2008). Multiple-Locus Sequence Typing Analysis of Bacillus thuringiensis Recovered from the Phylloplane of Clover (Trifolium hybridum) in Vegetative Form. Microbial Ecol 55: 619-625. Castillo JA, Greenberg JT (2007). Evolutionary dynamics of Ralstonia solanacearum. Appl Environ Microbiol 73: 1225-38. 11 Coscolla M, Gonzalez-Candelas F (2007). Population structure and recombination in environmental isolates of Legionella pneumophila. Environ Microbiol 9: 643-56. Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG (2004). Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol 42: 5644-9. Dalmastri C, Baldwin A, Tabacchioni S, Bevivino A, Mahenthiralingam E, Chiarini L et al (2007). Investigating Burkholderia cepacia complex populations recovered from Italian maize rhizosphere by multilocus sequence typing. Environ Microbiol 9: 1632-9. de Las Rivas B, Marcobal A, Munoz R (2004). Allelic diversity and population structure in Oenococcus oeni as determined from sequence analysis of housekeeping genes. Appl Environ Microbiol 70: 7210-9. de Las Rivas B, Marcobal A, Munoz R (2006). Development of a multilocus sequence typing method for analysis of Lactobacillus plantarum strains. Microbiol 152: 85-93. Diancourt L, Passet V, Chervaux C, Garault P, Smokvina T, Brisse S (2007). Multilocus Multilocus Sequence Typing of Lactobacillus casei (L. paracasei) reveals a clonal population structure with low levels of homologous recombination. Appl Environ Microbiol. 73: 66016611 Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43: 4178-82. 12 Diavatopoulos DA, Cummings CA, Schouls LM, Brinig MM, Relman DA, Mooi FR (2005). Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, humanassociated lineage of B. bronchiseptica. PLoS Pathog 1: e45. Edwards MT, Fry NK, Harrison TG (2008). Clonal population structure of Legionella pneumophila inferred from allelic profiling. Microbiol 154: 852-64. Enersen M, Olsen I, van Winkelhoff AJ, Caugant DA (2006). Multilocus sequence typing of Porphyromonas gingivalis strains from different geographic origins. J Clin Microbiol 44: 3541. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG (2000). Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38: 1008-15. Enright MC, Spratt BG, Kalia A, Cross JH, Bessen DE (2001). Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect Immun 69: 2416-27. Gonzalez-Escalona N, Martinez-Urtaza J, Romero J, Espejo RT, Jaykus LA, Depaola A (2008). Determination of Molecular Phylogenetics of Vibrio parahaemolyticus Strains by Multilocus Sequence Typing. J Bacteriol 190: 2831-2840 Goss EM, Kreitman M, Bergelson J (2005). Genetic diversity, recombination and cryptic clades in Pseudomonas viridiflava infecting natural populations of Arabidopsis thaliana. Genetics 169: 21-35. 13 Hanage WP, Kaijalainen T, Herva E, Saukkoriipi A, Syrjanen R, Spratt BG (2005). Using multilocus sequence data to define the pneumococcus. J Bacteriol 187: 6223-30. Homan WL, Tribe D, Poznanski S, Li M, Hogg G, Spalburg E et al (2002). Multilocus sequence typing scheme for Enterococcus faecium. J Clin Microbiol 40: 1963-71. Jiggins FM, von Der Schulenburg JH, Hurst GD, Majerus ME (2001). Recombination confounds interpretations of Wolbachia evolution. Proc Biol Sci 268: 1423-7. Jolley KA, Wilson DJ, Kriz P, McVean G, Maiden MC (2005). The influence of mutation, recombination, population history, and selection on patterns of genetic diversity in Neisseria meningitidis. Mol Biol Evol 22: 562-9. Lemee L, Dhalluin A, Pestel-Caron M, Lemeland JF, Pons JL (2004). Multilocus sequence typing analysis of human and animal Clostridium difficile isolates of various toxigenic types. J Clin Microbiol 42: 2609-17. Lodders N, Stackebrandt E, Nubel U (2005). Frequent genetic recombination in natural populations of the marine cyanobacterium Microcoleus chthonoplastes. Environ Microbiol 7: 434-42. Mayor D, Zeeh F, Frey J, Kuhnert P (2007). Diversity of Mycoplasma hyopneumoniae in pig farms revealed by direct molecular typing of clinical material. Vet Res 38: 391-8. Meats E, Feil EJ, Stringer S, Cody AJ, Goldstein R, Kroll JS et al (2003). Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol 41: 1623-36. 14 Miller SR, Castenholz RW, Pedersen D (2007). Phylogeography of the thermophilic cyanobacterium Mastigocladus laminosus. Appl Environ Microbiol 73: 4751-9. Nicolas P, Mondot S, Achaz G, Bouchenot C, Bernardet JF, Duchaud E (2008). Population structure of the fish-pathogenic bacterium Flavobacterium psychrophilum. Appl Environ Microbiol. Octavia S, Lan R (2006). Frequent recombination and low level of clonality within Salmonella enterica subspecies I. Microbiol 152: 1099-108. Olvera A, Cerda-Cuellar M, Aragon V (2006). Study of the population structure of Haemophilus parasuis by multilocus sequence typing. Microbiol 152: 3683-90. Pannekoek Y, Morelli G, Kusecek B, Morre SA, Ossewaarde JM, Langerak AA et al (2008). Multi locus sequence typing of Chlamydiales: clonal groupings within the obligate intracellular bacteria Chlamydia trachomatis. BMC Microbiol 8: 42. Papke RT, Koenig JE, Rodriguez-Valera F, Doolittle WF (2004). Frequent recombination in a saltern population of Halorubrum. Science 306: 1928-9. Perez-Losada M, Browne EB, Madsen A, Wirth T, Viscidi RP, Crandall KA (2006). Population genetics of microbial pathogens estimated from multilocus sequence typing (MLST) data. Infect Genet Evol 6: 97-112. Rasback T, Johansson KE, Jansson DS, Fellstrom C, Alikhani MY, La T et al (2007). Development of a multilocus sequence typing scheme for intestinal spirochaetes within the genus Brachyspira. Microbiol 153: 4074-87. 15 Ruiz-Garbajosa P, Bonten MJ, Robinson DA, Top J, Nallapareddy SR, Torres C et al (2006). Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J Clin Microbiol 44: 2220-8. Salcedo C, Arreaza L, Alcala B, de la Fuente L, Vazquez JA (2003). Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J Clin Microbiol 41: 757-62. Salerno A, Deletoile A, Lefevre M, Ciznar I, Krovacek K, Grimont P et al (2007). Recombining population structure of the Enterobacteriaceae Plesiomonas shigelloides revealed by multilocus sequence typing. J Bacteriol 189: 7808-18 Sarkar SF, Guttman DS (2004). Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl Environ Microbiol 70: 1999-2012. Silva C, Vinuesa P, Eguiarte LE, Souza V, Martinez-Romero E (2005). Evolutionary genetics and biogeographic structure of Rhizobium gallicum sensu lato, a widely distributed bacterial symbiont of diverse legumes. Mol Ecol 14: 4033-50. Sorokin A, Candelon B, Guilloux K, Galleron N, Wackerow-Kouzova N, Ehrlich SD et al (2006). Multiple-locus sequence typing analysis of Bacillus cereus and Bacillus thuringiensis reveals separate clustering and a distinct population structure of psychrotrophic strains. Appl Environ Microbiol 72: 1569-78. Stoddard RA, Miller WG, Foley JE, Lawrence J, Gulland FM, Conrad PA et al (2007). Campylobacter insulaenigrae isolates from northern elephant seals (Mirounga angustirostris) in California. Appl Environ Microbiol 73: 1729-35. 16 Tanabe Y, Kasai F, Watanabe MM (2007). Multilocus sequence typing (MLST) reveals high genetic diversity and clonal population structure of the toxic cyanobacterium Microcystis aeruginosa. Microbiol 153: 3695-3703. Thaipadungpanit J, Wuthiekanun V, Chierakul W, Smythe LD, Petkanchanapong W, Limpaiboon R et al (2007). A Dominant Clone of Leptospira interrogans Associated with an Outbreak of Human Leptospirosis in Thailand. PLoS Negl Trop Dis 1: e56. Vergin KL, Tripp HJ, Wilhelm LJ, Denver DR, Rappe MS, Giovannoni SJ (2007). High intraspecific recombination rate in a native population of Candidatus pelagibacter ubique (SAR11). Environ Microbiol 9: 2430-40. Vos M, Velicer GJ (2008). Isolation by distance in the spore-forming soil bacterium Myxococcus xanthus. Curr Biol 18: 386-91. Walk ST, Alm EW, Calhoun LM, Mladonicky JM, Whittam TS (2007). Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ Microbiol 9: 2274-88. Whitaker RJ, Grogan DW, Taylor JW (2005). Recombination shapes the natural population structure of the hyperthermophilic archaeon Sulfolobus islandicus. Mol Biol Evol 22: 235461. 17