Our goal is to develop artificial genetic switches or
advertisement

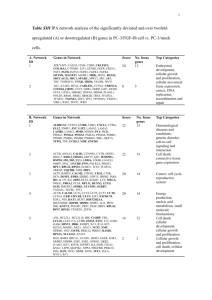
SUPPORTING INFORMATION Distinct DNA-based epigenetic switches trigger transcriptional activation of silent genes in human dermal fibroblasts Ganesh N. Pandiana+, Junichi Taniguchib+, Syed Junethab, Shinsuke Satoa, Le Hanb, Abhijit Sahab, Chandran AnandhaKumarb, Toshikazu Bandob, Hiroki Nagasec, d, Thangavel Vaijayanthib, Rhys D. Taylorb and Hiroshi Sugiyamaa, b*. 0 Methods General. Unless specified, all the reagents and solvents used in this work were purchased from standard suppliers and used without further purification. Abbreviations used: P, N-methylpyrrole; I, N-methylimidazole; AF, ammonium formate; Fmoc, 9-fluorenylmethoxycarbonyl;HCTU,1-[bis(dimethylamino)methylene]-5-chloro-1H-benzothiazolium 3-oxide hexafluorophosphate; DMSO, dimethylsulfoxide; DIEA, N,N-diisopropylethylamine; β, 1 H NMR spectra were recorded on a JEOL JNM-FX 400 model NMR spectrometer. HPLC analysis was performed with a 4.6 x 150 mm column on a JASCO PU-2089 plus pump model with UV-2075 plus HPLC UV/VIS detector and a Chemcobond 5-ODS-H 10 x 150 mm column (Chemco Scientific Co., Ltd, Osaka, Japan) was used for the purification of SAHA-PIPs and their precursor. Electrospray Ionization Time-of-Flight mass (ESI-TOF-MS) was recorded on BioTOF II ESI-TOF Bruker Daltonics Mass Spectrometer (Bremen, Germany). Flash column system was performed using Combi Flash Companion model (Teledyne Isco Inc., NE, USA). Teledyne Isco Redi Sep Rf column (40 g) was used for the purification of 4-(8-methoxy-8-oxooctanamido)benzoic acid. Synthesis and purification of SAHA-PIP conjugates Synthesis and characterization of 4-(8-methoxy-8-oxooctanamido) benzoic acid and subsequent 1 conversion to SAHA-PIP conjugates were performed on a PSSM-8 peptide synthesizer (Shimadzu, Kyoto) as mentioned before1,2. The purities of the all SAHA-PIPs (1-32) were checked by HPLC (elution with trifluoroacetic acid and a 0-100% acetonitrile linear gradient (0-40 min) at a flow rate of 1.0 mL min-1 under 254 nm). Cell culture HDF from 54-year-old Caucasian were purchased from Cell Applications, Inc. We used these cells for the screening studies within six passages. HDFs were maintained in Dulbecco's modified eagle medium (DMEM, Nacalai Tesque, Japan) containing 10% fetal bovine serum (FBS, Japan Serum). Cytotoxicity assay Colorimetric assays using WST-8 (Dojindo, Kumamoto, Japan) were carried out in 96-well plates with various concentrations of SAHA-PIPs as mentioned before23. Optimization of parameters for treatment of SAHA-PIP against HDF HDF cells with in the passage P6 were trypsinized for 5 min at 37 °C, and were resuspended in the fresh HDF medium to a concentration of 1.5 x 105 cells/ml in a 35 mm plate and were grown for 24 h as mentioned before3. The medium was then removed and replaced with 2 ml of fresh HDF medium followed by the addition of 1 mM of each individual SAHA-PIP (1 to 32) to achieve a final 2 polyamide concentration of 1 µM in 0.1% DMSO and then were incubated in a 5% CO2 atmosphere at 37 °C for 48 h. 0.1% DMSO treated cells were used as the control. The effective concentration of the SAHA-PIPs was standardized based on the initial optimization experiments and the treatment of HDF with various concentration of PI polyamide SAHA conjugates (100 nM, 500 nM, 1 µM and 10 µM)3. Studies with varied incubation time (24 h, 48 h and 72 h) suggested that 48 h is optimal to achieve consistent expression3. Hence, 1 µM of effectors and 48 h incubation was employed for all studies. 3 Figure S1 Figure S1. Number of genes affected with the treatment of SAHA, individual SAHA-PIPs 1-32 in HDFs for 48 h. a) up-regulated by more than 10-fold b) down-regulated by more than 10-fold. . Each bar is the summary of results derived from two individually treated culture plates 4 Figure S2 Figure S2. Heat map of top-100 down-regulated genes. An unsupervised hierarchical clustering analysis of top 100 down-regulated genes with the treatment of SAHA, SAHA-PIPs 1-32 is provided. Each result represents the summary of analysis of data derived from two culture plates 5 Figure S3 Figure S3. Functional annotation of the SAHA-PIP a) 1, b) 7 and c) 19 induced genes in human dermal fibroblasts. Dark gray bar indicates a particular set of biological processes that repeatedly occur. Each result is the summary of analysis of data derived from two individual culture plates. Data were analyzed through the use of IPA (Ingenuity® Systems, www.ingenuity.com). 6 Figure S4 7 8 9 Figure S4. Network analysis of SAHA-PIP a) 2, b) 13, c) 17, d) 18, e) 24 and f) 25 induced genes in human dermal fibroblasts corresponding to hematological system, nervous system, hair and skin, respiratory, sensory system and digestive system, respectively. Four-fold induction was chosen as the remarkable effect to account for the down-stream effect. Each result is the summary of analysis of data derived from two individual culture plates. Data were analyzed through the use of IPA (Ingenuity® Systems, www.ingenuity.com). 10 Figure S5 Figure S5. Heat map of SAHA-PIP induced ncRNAs. An unsupervised hierarchical clustering analysis of top 100 down regulated non-coding RNAs (ncRNAs) with the treatment of SAHA, SAHA-PIPs 1-32 is provided. Each result is the summary of analysis of data derived from two individual culture plates. 11 Figure S6 Figure S6. SAHA-PIPs are non-toxic to HDFs. Cytotoxicity studies carried out as mentioned before3 suggest that at a) 1 μM concentration SAHA but none of the SAHA-PIPs killed 50% of the cells. Even at b) 10 μM concentration none of the SAHA-PIPs were cytotoxic. Each bar represents mean ± SD from 9 well plates. 12 SUPPLEMENTARY TABLES Table S1. Number of up-/down-regulated genes in SAHA and SAHA-PIP (1-32)-treated HDFs. Compound Up-regulated Down-regulated >2 fold >5 fold >10 fold >2 fold >5 fold >10 fold 1 1331 656 515 927 85 61 2 1809 718 498 608 104 57 3 1399 499 289 524 124 65 4 703 225 153 992 96 61 5 1489 490 217 571 110 52 6 1314 314 156 683 115 59 7 3816 449 194 2414 219 58 8 3244 453 243 2314 195 69 9 2258 692 331 382 99 54 10 2406 257 131 1705 128 57 11 909 281 162 623 127 64 12 520 85 34 543 111 59 13 1468 693 433 626 91 45 14 1050 614 410 718 114 63 15 910 575 413 882 111 68 16 689 127 41 553 164 69 17 699 355 223 645 32 7 18 728 388 213 655 45 7 19 690 382 201 702 48 8 20 755 390 240 759 48 8 21 500 258 171 906 32 7 22 472 224 155 890 26 6 23 656 342 230 669 24 6 24 907 514 337 549 18 5 25 608 363 225 659 25 4 26 834 407 234 424 32 7 27 649 370 242 653 25 5 28 883 469 279 527 33 11 29 1201 117 12 330 69 14 30 681 80 12 334 53 11 31 870 87 5 318 42 12 32 288 38 4 243 30 8 SAHA 2002 281 47 1386 182 74 *Sum of data derived from two individual culture plates 13 Table S2. Summary of common genes up regulated by more than 10-fold between individual effector-treated HDFs. *Sum of data derived from two individual culture plates 14 Table S3. Summary of common genes down regulated by more than 10-fold between individual effector-treated HDFs. *Sum of data derived from two individual culture plates 15 Table S4. Ct values in qRT-PCR experiment SAHA-PIP Gene Ct - SAHA-PIP Ct - SAHA Ct - DMSO 1 GRPR 26.3 ± 1.2 39.6 ± 0.5 38.5 ± 2.0 CD24 16.7 ± 0.4 30.5 ± 0.0 31.1 ± 1.4 ACTB 12.9 ± 0.5 13.7 ± 0.7 13.5 ± 0.5 HLA-DOA 27.4 ± 2.1 38.9 ± 1.5 37.4 ± 0.3 DPYSL5 25.3 ± 1.1 40 ± 0 40 ± 0 ACTB 14.0 ± 0.8 14.9 ± 0.5 14.5 ± 0.9 GPC3 21.2 ± 0.1 30.1 ± 0.5 28.8 ± 0.0 SEMA6A 23.2 ± 0.0 29.0 ± 0.0 28.7 ± 0.6 ACTB 12.9 ± 0.1 13.7 ± 0.7 13.5 ± 0.5 PRSS8 22.3 ± 0.1 34.7 ± 0.2 34.6 ± 1.3 WNK2 24.5 ± 0.1 37.8 ± 3.0 36.7 ± 1.3 ACTB 13.0 ± 0.0 14.3 ± 0.8 14.1 ± 0.4 GPRC5B 27.2 ± 0.7 33.2 ± 0.1 32.5 ± 1.2 ACTB 14.2 ± 0.2 15.6 ± 0.1 15.1 ± 1.5 PDLIM3 22.5 ± 1.1 30.5 ± 0.0 32.4 ± 1.2 ACTB 11.8 ± 0.9 13.6 ± 0.1 13.8 ± 0.3 LEFTY1 19.2 ± 0.0 31.4 ± 1.1 30.0 ± 0.8 KSR2 31.9 ± 0.7 40 ± 0 40 ± 0 ACTB 12.0 ± 0.9 13.6 ± 0.1 13.8 ± 0.3 SMOC2 24.1 ± 0.8 38.1 ± 2.6 32.6 ± 0.8 TSTD1 21.9 ± 0.2 33.5 ± 1.0 32.5 ± 0.0 ACTB 13.8 ± 0.2 13.7 ± 0.1 13.9 ± 0.3 ATCAY 22.9 ± 0.2 30.0 ± 0.6 32.1 ± 0.7 ACTB 15.0 ± 0.0 14.0 ± 0.3 14.3 ± 0.2 STRA6 28.0 ± 1.2 39.6 ± 0.5 37.7 ± 3.2 ACTB 15.2 ± 1.5 16.1 ± 0.5 15.7 ± 1.0 MYO7A 31.1 ± 0.8 38.7 ± 0.8 37.5 ± 0.9 RBFOX3 25.2 ± 0.8 37.9 ± 2.5 33.2 ± 1.2 ACTB 14.8 ± 0.8 16.1 ± 0.5 15.7 ± 1.0 A_21_P0000813 22.3 ± 0.5 28.2 ± 0.2 30.9 ± 0.4 A_21_P0000821 25.2 ± 0.8 35.5 ± 1.3 36.9 ± 4.3 A_21_P0014207 24.3 ± 0.4 38.5 ± 2.0 39.5 ± 0.5 A_19_P00319154 25.1 ± 0.6 39.5 ± 0.6 40 ± 0 ACTB 13.2 ± 0.1 13.6 ± 0.1 13.2 ± 0.1 2 7 10 13 17 18 19 23 24 25 9 Mean ± SD from 6 wells and Ct cut-off value is 40. 16 Table S5. Primers for q-RT PCR analysis Genes Endogenous primers Genes Endogenous primers hACTB F: CAATGTGGCCGAGGACTTTG hKSR2 F: CTTCGAGGAGATGAACCTGT R: CATTCTCCTTAGAGAGAAGTGG hGRPR F: GGATCTCCCCGTGAACGATG R: AAGCGGCCCTTTCCAATGA hTSTD1 R: TGGAACGTTTCGCATGGACT hCD24 F: CAGTAGTCTTGATGACCAAAGTCC R: TTGCCCATCTGACAGAAGAAA hSMOC2 R: TCACACACACAGTAGCTTCAA hHLA-DOA F: TAACCGGCTCTGGATGACTC F: CTGTGTCAACCCCAAGACGA hATCAY F: GGAACGTTCATTCCCCGCTG hSYTL1 F: ATTGACGTAAGCGGCTGTCT hMYO7A F: CCTTTGTGCAGCTTCGAGGA hRBFOX3 F: ACCATTGTGCCAAATGCACC A_21_P0000813 F: GCATGTGCCAAAGAAGAGGC A_21_P0000821 F: CAGAAGGATTGTGCAGATTGT R: TTTCACATAGCAGGCATTTG hLEFTY1 F: GAACTCCCTGGACACTAGCAGAAT R: CCCTGACACTGCTTTATTTCTAACC A_21_P0014207 R: GGCCTGTGTTCTCACCATAGA hPDLIM3 F: TGCTGGGCCACAAGTTCA R: CGACAGTGTTGTCAGATTTTCTATTTAA R: AGGGGTGGCCCTCATATTCT hGPRC5B F: CTTTGTCCTCTGTACAGAGC R: TGAATGGTCACACCTTGG R: TTTCTGCCCTGTTACTCCCAC hWNK2 F: CATGAGTGATGGCAGTGAGAAG R: TCAGAGTCAAATGCACCAGC R: AAGGGTGGAAAGCAAAGGCA hPRSS8 F: GAGTCAGCCTTCTTCCCTCACG R: GGTCCAGTATGGGAGGACACC R: GTCAGTGCACCAGGAAGAAGA hSEMA6A F: GGTTTCCCTCTGGTGACACAT R: CCAAAGCTCATGGAGACGA R: CTGCACCAGAACACAACGTG hGPC3 F: CTGAGACATGCCTTGTAGAAT R: TGTGGCTCCAAAGGCTTGAT R: TGGCTGATGCCCTAACAGAC hDPYSL5 F: TTGGAGAGTGCTCTGCAGAT F: GGTTGTACTCTGGCCATTAATGTG R: CGATTACCCAAGACCCCAGAA A_19_P00319154 F: CAGCGTACAAACCTGCATAAAAGA R: CACCCGGTTGAATCAAATCC F: TGTGGAGATACTGTAACCTGAG R: ATGCACAACCCACACTTAACC 17 References 1. Pandian, G. N. et al. Synthetic small molecules for epigenetic activation of pluripotency genes in mouse embryonic fibroblasts. ChemBioChem 12, 2822-2828, (2011). 2. Pandian, G. N. et al. A synthetic small molecule for rapid induction of multiple pluripotency genes in mouse embryonic fibroblasts. Sci. Rep. 2, e544(2012). 3. Han, L. et al. A synthetic small molecule for targeted transcriptional activation of germ cell genes in a human somatic cell. Angew. Chem. Int. Ed., 52, 13410 –13413, (2013). . 18