jcb_22175_sm_SupplTabS3
advertisement

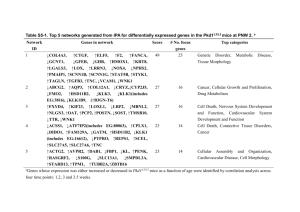
1 Table SIII IPA network analysis of the significantly deviated and over twofold upregulated (A) or downregulated (B) genes in PC-3/FGF-8b cell vs. PC-3/mock cells. A. Network ID 1 Genes in Network Score ADCYAP1, CALD1, CD44, CDH5, CKLFSF3, COL18A1, CTNNB1, E2F1, EFNB2, EGFR, FBLN2, FGF3, FGF8, FGF10, FGFR1, FGFR3, FGFR4, IFITM1, MAGEF1, MAPK1, MDK, MSX1, RGS12, SDCCAG33, SHC1, SPARC, SPRY2, SRC, SRF, TNF, TNFRSF21, TNXB, TRIP6, VEGFB, WNT1 AK1, ATAD2, BTG2, CABLES1, CCNG1, CDKN2A, CITED4, CRIP1, CTCF, DDR1, DHFR, E4F1, EP300, FEN1, GTSE1, HMGB2, HNF4A, MDM4, PLAGL1, POLD3, RRM1, RRM2, RRM2B, TBX3, TFAP2A, TFAP2C, TM7SF2, TOP1, TP53, TP53INP1, TWIST1, VRK1, WIG1, WNT1, WRN 24 No. focus genes 12 8 5 B. Network ID 1 Genes in Network Score ALDH1A3, ATXN3, CCRK, CDK2, CXCL1, CYBA, ELL2, FNBP1, FST, G1P2, LAMA2, LAMA3, LAMB3, LAMC2, MMD, NEDD8, PC4, PIGR, PSMA1, PSMA2, PSMA3, PSMA4, PSMA6, PSMB1, PSMB2, PSMB6, PSMB8, PSMB10, SIM1, SKP1A, TFPI2, TNF, UCHL3, VDR, ZNF365 22 No. focus genes 15 2 ACTB, ANXA2, CALB1, CTNNB1, CYTB, DDIT3, DDX17, EDIL3, FBXO2, GDF15, GH1, HNRPH1, HSPD1, ID1, ID3, IFI16, LDHA, LDHB, LGALS3, MMP7, MYC, MYCBP, PNN, POLR2A, PRDX3, RPL7, RPL22, RPS23, SUMO1, TCF4, TFAP2A, TRRAP, UQCRH, XDH, ZRF1 ACP5, BARD1, CALM2, CITED2, CKS2, CTSK, DCN, DUSP1, EPRS, ERBB2, GDF15, HSPB2, IARS, ID1, IL17F, IL6, JDP2, KLF4, KLKB1, LYZ, MIG-6, P4HA1, P4HA2, PCAF, RPL11, RUNX3, RXRB, SLPI, SMURF2, SPHK1, STATIP1, SURB7, TADA3L, TGFB1, TP53 ACTB, CALM2, CCT3, CCT4, CCT5, CCT7, CCT8, CCT6A, CKB, CRYAB, CYCS, E2F1, EXOSC10, FDX1, FOS, HAT1, HCRT, HIST2H2AA, HIST2H2BE, HSPB1, HSPD1, HSPE1, IL15, IPO9, JRK, KNTC2, PMAIP1, PRF1, PSAP, RBP1, RPL26, RPS7, RPS24, TXNRD1, ZFP36 22 15 20 14 Cancer, cell cycle, reproductive system 20 14 AXL, BCL2L1, BCL2L10, BIK, C1QBP, CBL, CCL26, CLEC11A, CYBB, E2IG5, ETF1, F12, GAB1, GAS6, HINT1, HRK, IGFBP3, IL13, IL18, KIT, KITLG, MAPK1, MCL1, MUC1, NCF2, NMU, OPRM1, OXT, OXTR, PRKCQ, PRKD1, RASD1, RPS3A, SLC43A3, STAT6 BAX, BMP4, BRCA1, CCND1, DDIT3, EGFR, EGR1, ERBB3, ERBB4, ESR1, ESR2, GNRH1, GRB2, ICAM1, IGF1, IGF1R, IGFBP3, IL8, INSR, ITGA5, KNG1, LEPR, MAP3K1, NPPA, PDGFRB, PRKCD, PTH, SLPI, SNX1, SNX2, SNX6, TFF1, TGFA, WNT1, WT1 16 12 2 3 Energy production, nucleic acid metabolism, small molecule biochemistry Cell death, cellular development, cellular growth and proliferation Cellular growth and proliferation, cell death, cellular development 2 3 4 5 6 Top Categories Embryonal development, cellular growth and proliferation, cellular movement Gene expression, cancer, DNA replication, recombination and repair Top Categories Dermatological diseases and conditions, genetic disorder, cell-to-cell signaling and interaction Cell death, connective tissue gene expression 2 Table SIII footnote: Top networks and their top three network function categories generated by IPA for differentially (over twofold changed) expressed genes in FGF-8transfected PC-3 cells. The score is the negative log of p and indicates the likelihood of the focus genes in a network being found together as a result of chance. Therefore, scores of 2 have at least 99% confidence of not being generated by chance alone. No. focus genes refers to the genes affected by FGF-8 transfection within a network. Bold: genes affected by FGF-8; standard text: genes in network not affected by FGF-8.