Disulfide-bonded Self-assembling Protein Scaffold for an Artificial
advertisement

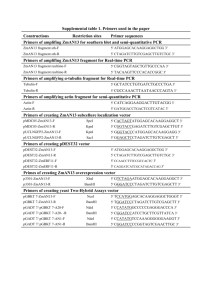
Supporting information Materials. Synthetic genes encoding PCNA1, PCNA2 and PCNA3 were purchased from GenScript (Piscataway, NJ, USA). pHSG398 was purchased from Takara Bio (Shiga, Japan). pET-15b(+), pET-28b(+) and pLysS were purchased from Novagen (Darmstadt, Germany). Anhydrotetracycline and pASK-IBA3 plus were purchased from IBA (Goettingen, Germany). pQE80L-Kan was purchased from Qiagen (Hilden, Germany). pBAD202/D/lacZ was purchased from Invitrogen (Carlsbad, CA, USA). T7 polynucleotide kinase, T4 DNA ligase and Escherichia coli T7 Express Iq were purchased from New England Biolabs (Ipswich, MA, USA). In-Fusion enzyme was purchased from Clontech Laboratories (Mountain View, CA, USA). Ampicillin, kanamycin, isopropyl β-D-1-thiogalactopyranoside (IPTG) and arabinose were purchased from Wako Pure Chemical Industries (Osaka, Japan). 5-Aminolevulinic acid hydrochloride (ALA) was purchased from COSMO BIO (Tokyo, Japan). HisTrap FF crude column (1.6×2.5 cm), HiTrap Q FF column (1.6×2.5 cm), HiTrap DEAE FF column (1.6×2.5 cm), HiLoad 16/600 Superdex 75 pg column (1.6×60 cm) and Superdex 200 10/300 GL (1.0×30 cm) were purchased from GE Healthcare (Little Chalfont, Buckinghamshire, UK). DE52 preswollen microgranular DEAE cellulose was purchased from Whatman (Maidstone, Kent, UK). Vector Construction. The gene encoding PCNA1 was amplified by PCR using two primers 5’-GGAATTCATATGTTTAAAATTGTGTATCCGAACGCC-3’ (forward) and 5’-CGGGATCC GCTCTTCACAGGCGCGGCGCAATCC-3’ (reverse). The generated DNA fragment was cloned into pHSG398 between EcoRI and BamHI sites. After the resulting plasmid was digested with 1 NdeI and BamHI, the generated fragment was ligated into pET-15b(+) that was digested with the same restriction enzymes. The resulting plasmid, pET15b+PCNA1WT, expresses PCNA1WT. The G108C mutation was introduced by PCR using two primers 5’-AAATCTTGCGCCAAAAGTAC CATCTAC-3’ (forward) and 5’-TTTGGCGCAAGATTTTTCATCACGGAT-3’ (reverse). The P186C mutation was introduced by PCR using two primers 5’-GATAAATGCCTGAAAGAACT GAGCATC-3’ (forward) and 5’-TTTCAGGCATTTATCTT TCATCAGAAA-3’ (reverse). The gene encoding PCNA2 was amplified by PCR using two primers 5’-GGAATTCAT ATGATGAAAGCCAAAGTGATCGATG-3’ (forward) and 5’-CGGGATCCGCTCTTCAATCC GCGCGCGGTGC-3’ (reverse). The generated DNA fragment was digested with NdeI and BamHI and ligated into pET-15b(+) that was digested with the same restriction enzymes. The resulting plasmid, pET15b+PCNA2WT, expresses PCNA2WT. The E105C mutation was introduced by PCR using two primers 5’-GATGGTTGCTTTACCCGCAGTTTTGAA-3’ (forward) and 5’-GGTAAAGCAACCATCAAACGTCAGGGT-3’ (reverse). The L171C mutation was introduced by PCR using two primers 5’-GGTGATTGCAGCACCGCGAAAGTT GAA-3’ (forward) and 5’-GGTGCTGCAATCACCGATCACTTCGAA-3’ (reverse). An expression vector for PCNA3WT, pET15b+PCNA3WT, was constructed as described for the construction of pET15b+PCNA1WT, except that two primers 5’-GGAATTCATATGATCT ACCTGAAATCTTTCGAACG-3’ (forward) and 5’-CGGGATCCGCTCTTCACACTTTCGGC GCCAGCAG-3’ (reverse) were used to amplify the PCNA3 encoding gene. The R112C mutation was introduced by PCR using two primers 5’-AACGTTTGCAATCTGGAAGTGTCTGAA-3’ (forward) and 5’-CAGATTGCAAACGTTAAATTCACGAT T-3’ (reverse). The T180C mutation was introduced by PCR using two primers 5’-AAAGATTGCGGCGGTCTGCAGGATCTG-3’ 2 (forward) and 5’-ACCGCCGCAATCTTTAGAAAATTCCAC-3’ (reverse). We modified pET-15b(+) and pASK-IBA3 plus in advance to constructed an expression plasmid for the PCNA3-P450cam fusion protein. pET-15b(+) was linearized by PCR using two primers 5’-ATTCGAACGCCAGCACATGGACAGCGGCAGCAGCGGCCTGGTGC-3’ (forward) and 5’-AGCAGCGGTTTCTTTGCCCATGGTATATCTCCTTCTTAAAGTTAAAC-3’ (reverse). The linearized plasmid was phosphorylated by T7 polynucleotide kinase and self-ligated by T4 DNA ligase. The resulting plasmid, pStag, has an N-terminal S•Tag sequence. pASK-IBA3 plus was linearized by PCR using two primers 5’-ATGGGAATTCAGCGCTTGGA GCCACC-3’ (forward) and 5’-GCCATGGTATATCTCCTTCTTAAAGTTAAACAAAATTATTT C-3’ (reverse) to introduce the NcoI and EcoRI sites. The linearized plasmid was phosphorylated by T7 polynucleotide kinase and self-ligated by T4 DNA ligase. The resulting plasmid was linearized again by PCR using two primers 5’-GGATCCTGTTTAAACGACCTGTGAAGTGAA AAATGGCG-3’ (forward) and 5’-GCTTATTCGAACTGCGGGTGGCTCCAAG-3’ (reverse) and fused with t0 terminator, which was amplified from pQE80L-Kan by PCR using two primers 5’-GCAGTTCGAATAAGCTTAATTAGCTGAGCTTGGACTC-3’ (forward) and 5’-GTTTAAA CAGGATCCGGATTCTCACCAATAAAAAACGCCC-3’ (reverse), by In-Fusion enzyme. The resulting plasmid, pASKt, was linearized by PCR using two primers 5’-GCCTTTTTACGGTTC CTGGC-3’ (forward) and 5’-TTTCTACGGGGTCTGACGC-3’ (reverse) and fused with a p15A replicon sequence, which was amplified from pLysS using two primers 5’- CAGACCCCGTAGAAAAGCGCTAGCGGAGTGTATAC-3’ (forward) and 5’-GAACCGTAAA AAGGCAGAATTACAACTTATATCGTATGGGG-3’ (reverse), by In-Fusion enzyme. The resulting plasmid, pASKt15, has a p15A replicon. 3 The gene encoding P450cam was amplified by PCR using two primers 5’-GGAATTCG CTCTTCAGTTGGCGGTAGCATGACGACTGAAACCATACAAAGCAAC-3’ (forward) and 5’-CGGGATCCTTATACCGCTTTGGTAGTCGCC-3’ (reverse). The generated DNA fragment was cloned into pHSG398 between the EcoRI and BamHI sites. The PmeI site was inserted after the stop codon by PCR using two primers 5’-TAAGTTTAAACGGATCCTCTAGAGTCGAC-3’ (forward) and 5’-ATCCGTTTAAACTTATACCGCTTTGGTAGT-3’ (reverse) to obtain plasmid pHSG+GS-P450cam. The gene encoding PCNA3, which was amplified by PCR using two primers 5’-GGAATTCATATGATCTACCTGAAATCTTTCGAACG-3’ (forward) and 5’-CGGG ATCCGCTCTTCACACTTTCGGCGCCAGCAG-3’ pHSG+GS-P450cam between the EcoRI and (reverse), SapI sites. was The cloned resulting into plasmid, pHSG+PCNA3-P450cam, was digested with NdeI amd BamHI, and the generated fragment was cloned into pStag that was digested with the same restriction enzymes. The resulting plasmid, pStag+P3C, was digested with NcoI and PmeI, and the generated fragment was cloned into pASKt15 that was digested with the same restriction enzymes. The resulting plasmid, pASKt15+P3C, expresses PCNA3WT-P450cam. The R112C and T180C mutations were introduced as described above. The gene encoding PCNA1 was amplified by PCR using two primers 5’-CGGGATCCA TATGTTTAAAATTGTGTATCCGAACGCC-3’ (forward) and 5’-GGAATTCGCTCTTCACAG GCGCGGCGCAATCC-3’ (reverse). The generated DNA fragment was cloned into pHSG398 between the BamHI and EcoRI sites to generate the plasmid pHSG+PCNA1. The gene encoding PdR, which was amplified from pT1R[S1] by PCR using two primers 5’-CGGGATCCGCTCTTC GCTGGGTGGCGGCGGTAGC-3’ (forward) and 5’-GGAATTCTCAGGCACTACTCAGTTCA 4 GC-3’ (reverse), was digested with EcoRI and SapI, and cloned into pHSG+PCNA1, which was digested with the same restriction enzymes. The resulting plasmid, pHSG+PCNA1-PdR, was digested with NdeI and BamHI. The generated DNA fragment was cloned into pET-28b(+) between the NdeI and BamHI sites. The resulting plasmid, pET28b+P1R, was digested with NcoI and EcoRI, and the generated fragment was cloned into pASKt that was digested with the same restriction enzymes. After the resulting plasmid, pASKt+P1R, was digested with NcoI and PmeI, the generated fragment was ligated into pBAD202/D/lacZ that was digested with the same restriction enzymes. The resulting plasmid, pBAD+P1R, expresses PCNA1WT-PdR. The G106C mutation was introduced as described above. The gene encoding the C73S/C85S mutant of PdX was amplified from pEX[S2] by PCR using two primers 5’-GGAATTCCTGGTGCCGCGCGGCAGCGGCGGTGGTGGCTCTATGT CTAAAGTAGTGTATGTGTCAC-3’ (forward) and 5’-GCTAGTTATTGCTCAGCGG-3’ (reverse). The generated fragment was further amplified by PCR using two primers 5’-GGAATT CGCTCTTCAGACGGCGGTGGCGGTAGCCTGGTGCCGCGCGGC-3’ (forward) and 5’-GCT AGTTATTGCTCAGCGG-3’ (reverse). The generated fragment was cloned into pHSG398 between the EcoRI and BamHI sites. The PmeI site was inserted after the stop codon by PCR using two primers 5’-TAAGTTTAAACGGATCCTCTAGAGTCGAC-3’ (forward) and 5’-ATCC GTTTAAACTTACCATTGCCTATCGGG-3’ (reverse) to obtain the plasmid pHSG+GStGS-PdX. The gene encoding PCNA2, which was amplified by PCR using two primers 5’-GGAATTCATA TGATGAAAGCCAAAGTGATCGATG-3’ (forward) and 5’-CGGGATCCGCTCTTCAGTCCG CGCGCGGTGC-3’ (reverse), was cloned into pHSG+GStGS-PdX between the EcoRI and SapI sites. The resulting plasmid, pHSG+PCNA2-PdX, was digested with NdeI and BamHI. The 5 generated fragment was cloned into pET-15b(+) that was digested with the same restriction enzymes. The resulting plasmid, pET15b+P2X, expresses PCNA2WT-PdX. The L171C mutation was introduced as described above. Protein Expression and Purification. E. coli T7 Express Iq was transformed with pET15b+PCNA1WT. A single colony of cells was inoculated into 5 mL LB medium containing 100 mg/L ampicillin and the cells were grown at 37°C until the OD at 600 nm (OD600) reached 1.0. The culture was added to 250 mL TB medium containing 100 mg/L ampicillin and the cells were grown at 37°C. After the OD600 reached about 0.5, 0.25 mmol of IPTG was added and the culture was continued at 27°C overnight. The cells were harvested by centrifugation at 6,000×g for 20 min and resuspended in 20 mM potassium phosphate buffer, pH 7.4, containing 150 mM KCl, 10 mM imidazole and 1 mM DTT. After the cells were disrupted by ultrasonication, cell debris was removed by centrifugation at 22,000×g for 30 min. The resulting cell lysate was loaded on a HisTrap FF crude column. After washing the column with the same buffer, PCNA1WT was eluted with a linear gradient of imidazole (10-300 mM). The eluted protein was diluted 7 times with 20 mM potassium phosphate buffer, pH 7.4, containing 1 mM EDTA and 5 mM DTT. The diluted protein was loaded on a HiTrap Q FF column. After washing the column with the above buffer, the protein was eluted with a linear gradient of KCl (0-500 mM). The eluted protein was concentrated with an Amicon Ultra-15 Centrifugal Unit (10,000 NMWL). The concentrated protein was subjected to size-exclusion chromatography on a HiLoad 16/600 Superdex 75 pg column with 50 mM potassium phosphate buffer, pH7.4, containing 150 mM KCl, 1 mM EDTA and 10 mM DTT. The purified PCNA1WT was concentrated with the above 6 centrifugal unit. PCNA1G108C, PCNA1P186C and PCNA1G108C/P186C were expressed and purified as described above. PCNA2WT, PCNA2E105C, PCNA2L171C, PCNA2E105C/L171C, PCNA3WT, PCNA3R112C, PCNA3T180C and PCNA3R112C/T180C were expressed and purified as described for PCNA1WT, except that the eluted protein from the HisTrap column was loaded on a HiTrap Q FF column without dilution and eluted with a linear gradient of KCl (150-500 mM). The purification yields of PCNA1WT, PCNA1G108C, PCNA1P186C, PCNA1G108C/P186C, PCNA2WT, PCNA2E105C, PCNA2L171C, PCNA2E105C/L171C, PCNA3WT, PCNA3R112C, PCNA3T180C and PCNA3R112C/T180C were 9.5, 6.3, 5.9, 15, 43, 47, 43, 13, 11, 7.2, 8.1 and 10 mg, respectively. E. coli T7 Express Iq was transformed with pBAD+P1R. A single colony of cells was inoculated into 5 mL LB medium containing 50 mg/L kanamycin and the cells were grown at 37°C until the OD600 reached 1.0. The culture was added to 1 L TB medium containing 50 mg/L kanamycin and the cells were grown at 37°C. After the OD600 reached about 0.5, 200 mg of arabinose was added and the culture was continued at 27°C overnight. The cells were harvested by centrifugation at 6,000×g for 20 min and resuspended in 20 mM potassium phosphate buffer, pH 7.4, containing 150 mM KCl, 10 mM imidazole and 1 mM DTT. After the cells were disrupted by ultrasonication, cell debris was removed by centrifugation at 22,000×g for 30 min. The resulting cell lysate was loaded on a HisTrap FF crude column. After washing the column with the same buffer, PCNA1WT-PdR was eluted with a linear gradient of imidazole (10-300 mM). The eluted protein was diluted 3 times with 20 mM potassium phosphate buffer, pH 7.4, containing and 5 mM DTT. The diluted protein was loaded on a HiTrap Q FF column. After washing the column with 20 mM potassium phosphate buffer, pH 7.4, containing 50 mM KCl 7 and 5 mM DTT, the protein was eluted with a linear gradient of KCl (50-500 mM). The fractions with a ratio of A280/A455 less than 10 were combined and concentrated with an Amicon Ultra-15 Centrifugal Unit (50,000 NMWL). The concentrated protein was subjected to size-exclusion chromatography on a HiLoad 16/600 Superdex 200 pg column with 50 mM potassium phosphate buffer, pH7.4, containing 150 mM KCl and 10 mM DTT. The fractions with a ratio of A280/A455 less than 7.9 were combined and concentrated with the above centrifugal unit. PCNA1G108C-PdR was expressed and purified as described above. The concentrations of PCNA1WT-PdR and PCNA1G108C-PdR were calculated using 455 = 11.0 mM-1 cm-1.[S1] PCNA2WT-PdX and PCNA2L171C-PdX were expressed and purified as described for PCNA2WT, except that cells were cultured in 1 L TB medium, protein expression was induced by 1 mmol of IPTG, buffers did not contain EDTA, the fractions with a ratio of A412/A280 higher than 0.25 were collected after the anion exchange chromatography, size-exclusion chromatography was performed on a HiLoad 16/600 Superdex 200 pg column and the fractions with a ratio of A412/A280 higher than 0.30 were collected after the size-exclusion chromatography. The concentrations of PCNA2WT-PdX and PCNA2L171C-PdX were calculated using 412 = 11.0 mM-1 cm-1.[S3] E. coli T7 Express Iq was transformed with pASKt15+P3C. A single colony of cells was inoculated into 5 mL LB medium containing 100 mg/L ampicillin and the cells were grown at 37°C until the OD600 reached 1.0. The culture was added to 1 L TB medium containing 100 mg/L ampicillin and the cells were grown at 37°C. After the OD600 reached about 0.5, 200 g of anhydrotetracycline and 1 mmol of ALA were added and the culture was continued at 27°C overnight. The cells were harvested by centrifugation at 6,000×g for 20 min and resuspended in 8 20 mM potassium phosphate buffer, pH 7.4, containing 5 mM DTT and 5 mM d-camphor. After the cells were disrupted by ultrasonication, cell debris was removed by centrifugation at 22,000×g for 30 min. The supernatant was subjected to ammonium sulfate fractionation. PCNA3WT-P450cam precipitated at 30% saturation. The precipitated protein was dissolved in 20 mM potassium phosphate buffer, pH 7.4, containing 5 mM DTT and 5 mM d-camphor and dialyzed against the same buffer. The dialyzed protein was loaded on a DE52 column (2.5×8.5 cm). After washing the column with 20 mM potassium phosphate buffer, pH 7.4, containing 150 mM KCl, 5 mM DTT and 5 mM d-camphor, the protein was eluted with a linear gradient of KCl (150-500 mM). The red fractions were combined and diluted 2 times with 20 mM potassium phosphate buffer, pH 7.4, containing 5 mM DTT and 5 mM d-camphor. The diluted protein was loaded on a HiTrap Q FF column. After washing the column with 20 mM potassium phosphate buffer, pH 7.4, containing 150 mM KCl, 5 mM DTT and 5 mM d-camphor, the protein was eluted with a linear gradient of KCl (150-500 mM KCl). The fractions with a ratio of A392/A280 higher than 0.75 were combined and diluted 2 times with 50 mM potassium phosphate buffer, pH 7.4, containing 5 mM DTT and 5 mM d-camphor. After washing the column with the above buffer, the protein was eluted with a linear gradient of potassium phosphate (50-500 mM). The fractions with a ratio of A392/A280 higher than 1.0 were combined and concentrated with an Amicon Ultra-15 Centrifugal Unit (50,000 NMWL). The concentrated protein was subjected to size-exclusion chromatography on a HiLoad 16/600 Superdex 200 pg column with 50 mM potassium phosphate buffer, pH7.4, containing 150 mM KCl, 10 mM DTT and 5 mM d-camphor. The fractions with a ratio of A392/A280 higher than 1.1 were combined and concentrated with the above centrifugal unit. The concentrations 9 of PCNA3WT-P450cam and PCNA3R112C/T180C-P450cam were calculated using 392 = 90.2 mM-1 cm-1.[S1] 10 Protein Sequences PCNA1WT-PdR MGSSHHHHHHSSGLVPRGSHMFKIVYPNAKDFFSFINSITNVTDSIILNFTEDGIFSRHLTEDKV LMAIMRIPKDVLSEYSIDSPTSVKLDVSSVKKILSKASSKKATIELTETDSGLKIIIRDEKSGAK STIYIKAEKGQVEQLTEPKVNLAVNFTTDESVLNVIAADVTLVGEEMRISTEEDKIKIEAGEEGK RYVAFLMKDKPLKELSIDTSASSSYSAEMFKDAVKGLRGFSAPTMVSFGENLPMKIDVEAVSGGH MIFWIAPRLGGGGSGGGGSMNANDNVVIVGTGLAGVEVAFGLRASGWEGNIRLVGDATVIPHHLP PLSKAYLAGKATAESLYLRTPDAYAAQNIQLLGGTQVTAINRDRQQVILSDGRALDYDRLVLATG GRPRPLPVASGAVGKANNFRYLRTLEDAECIRRQLIADNRLVVIGGGYIGLEVAATAIKANMHVT LLDTAARVLERVTAPPVSAFYEHLHREAGVDIRTGTQVCGFEMSTDQQKVTAVLCEDGTRLPADL VIAGIGLIPNCELASAAGLQVDNGIVINEHMQTSDPLIMAVGDCARFHSQLYDRWVRIESVPNAL EQARKIAAILCGKVPRDEAAPWFWSDQYEIGLKMVGLSEGYDRIIVRGSLAQPDFSVFYLQGDRV LAVDTVNRPVEFNQSKQIITDRLPVEPNLLGDESVPLKEIIAAAKAELSSA PCNA2WT-PdX MGSSHHHHHHSSGLVPRGSHMMKAKVIDAVSFSYILRTVGDFLSEANFIVTKEGIRVSGIDPSRV VFLDIFLPSSYFEGFEVSQEKEIIGFKLEDVNDILKRVLKDDTLILSSNESKLTLTFDGEFTRSF ELPLIQVESTQPPSVNLEFPFKAQLLTITFADIIDELSDLGEVLNIHSKENKLYFEVIGDLSTAK VELSTDNGTLLEASGADVSSSYGMEYVANTTKMRRASDSMELYFGSQIPLKLRFKLPQEGYGDFY IAPRADGGGGSLVPRGSGGGGSMSKVVYVSHDGTRRELDVADGVSLMQAAVSNGIYDIVGDCGGS ASCATCHVYVNEAFTDKVPAANEREIGMLESVTAELKPNSRLSCQIIMTPELDGIVVDVPDRQW PCNA3WT-P450cam MGKETAAAKFERQHMDSGSSGLVPRGSHMIYLKSFERNIRLINMKVVYDDVRVLKDIIQALARLV DEAVLKFKQDSVELVALDRAHISLISVNLPREMFKEYDVNDEFKFGFNTQYLMKILKVAKRKEAI EIASESPDSVIINIIGSTNREFNVRNLEVSEQEIPEINLQFDISATISSDGFKSAISEVSTVTDN VVVEGHEDRILIKAEGESEVEVEFSKDTGGLQDLEFSKESKNSYSAEYLDDVLSLTKLSDYVKIS FGNQKPLQLFFNMEGGGKVTYLLAPKVGGSMTTETIQSNANLAPLPPHVPEHLVFDFDMYNPSNL SAGVQEAWAVLQESNVPDLVWTRCNGGHWIATRGQLIREAYEDYRHFSSECPFIPREAGEAYDFI PTSMDPPEQRQFRALANQVVGMPVVDKLENRIQELACSLIESLRPQGQCNFTEDYAEPFPIRIFM LLAGLPEEDIPHLKYLTDQMTRPDGSMTFAEAKEALYDYLIPIIEQRRQKPGTDAISIVANGQVN GRPITSDEAKRMCGLLLVGGLDTVVNFLSFSMEFLAKSPEHRQELIERPERIPAACEELLRRFSL VADGRILTSDYEFHGVQLKKGDQILLPQMLSGLDERENACPMHVDFSRQKVSHTTFGHGSHLCLG QHLARREIIVTLKEWLTRIPDFSIAPGAQIQHKSGIVSGVQALPLVWDPATTKA 11 Table Table S1. Distances between alpha-carbons in disulfide bonds listed in the previous report.[S4] PDB ID 1A8E 1ABA 1AHO 1B3A 1BEB 1BQC 1C2A 1CNV 1D0D Cysteines Distance (Å) 9C-48C 19C-39C 118C-194C 137C-331C 158C-174C 161C-179C 171C-177C 227C-241C 14C-17C 12C-63C 22C-46C 26C-48C 10C-34C 11C-50C 66C-160C 106C-119C 74C-81C 9C-63C 10C-25C 15C-23C 32C-39C 36C-51C 68C-122C 69C-84C 74C-82C 91C-98C 95C-110C 23C-72C 41C-93C 54C-62C 5C-55C 14C-38C 30C-51C 5.8 6.1 6.0 6.0 5.2 4.6 5.1 4.6 5.2 4.9 5.3 5.6 5.6 6.0 6.1 3.8 5.7 5.2 6.1 4.1 6.3 6.0 5.0 6.1 4.0 6.3 6.2 5.6 5.2 4.9 5.8 5.7 6.4 PDB ID 1DY5 1E5P 1EDM 1EN2 1EZM 1H03 1HX0 1HXN 1I0V 12 Cysteines Distance (Å) 26C-84C 40C-95C 58C-110C 65C-72C 38C-42C 57C-149C 51C-62C 56C-71C 73C-82C 3C-18C 12C-24C 17C-31C 35C-39C 49C-64C 58C-70C 63C-75C 82C-86C 30C-58C 270C-297C 7C-48C 34C-64C 69C-111C 97C-127C 28C-86C 70C-115C 141C-160C 378C-384C 450C-462C 229C-432C 338C-380C 390C-407C 2C-10C 6C-103C 5.6 5.5 5.9 5.3 4.6 5.7 5.8 5.6 5.4 6.0 5.8 5.2 5.9 6.0 5.7 5.3 6.0 5.7 4.8 6.0 5.9 5.9 5.5 5.3 5.6 5.1 6.2 5.3 5.7 4.9 6.2 5.3 4.5 PDB ID 1I4U 1I71 1I8E 1JND 1JU2 1K07 1K5C 1K5N 1ME4 1MSO 1QDD 1QFO 1QNR Cysteines Distance (Å) 12C-121C 57C-173C 117C-150C 1C-78C 22C-61C 50C-73C 5C-17C 12C-30C 24C-41C 6C-33C 322C-405C 399C-450C 256C-290C 3C-17C 175C-191C 300C-303C 25C-80C 101C-164C 203C-259C 22C-63C 56C-95C 153C-200C 6C-11C 7C-7C* 19C-20C* 14C-25C 42C-140C 115C-132C 22C-79C 26C-29C 172C-175C 265C-272C 284C-334C 6.1 5.6 5.4 4.9 5.7 6.3 5.8 5.8 5.2 5.9 5.8 6.4 6.4 5.9 5.8 5.9 6.3 6.3 6.4 5.4 4.9 5.4 4.9 4.6 5.3 5.9 5.5 5.1 4.8 5.6 5.4 5.6 6.0 PDB ID 1QQ4 1SGP 1TML 2DNJ 2MCM 2MSB 2PSP 2TGI 3LZT 3SEB 3SIL *Intersubunit disulfide bond 13 Cysteines Distance (Å) 17C-37C 101C-111C 137C-170C 8C-38C 16C-35C 24C-56C 42C-58C 191C-220C 80C-125C 232C-267C 173C-209C 36C-46C 88C-93C 128C-217C 195C-209C 6C-104C 8C-35C 19C-34C 29C-44C 58C-84C 68C-83C 78C-95C 7C-16C 15C-78C 44C-109C 48C-111C 6C-127C 30C-115C 64C-80C 76C-94C 93C-113C 42C-103C 6.3 3.9 4.9 5.2 5.7 4.7 6.5 5.1 5.7 6.1 5.8 3.9 5.6 5.4 5.0 4.9 5.7 6.6 5.9 5.7 6.5 5.9 5.7 5.3 5.8 5.9 5.0 6.1 4.9 5.3 5.5 6.4 Figures Figure S1. Histogram of distance between alpha-carbons in disulfide bond. Figure S2. Elution profiles of the mixtures of i) PCNA1WT, PCNA2WT and PCNA3WT (black), ii) PCNA1P186C, PCNA2E105C and PCNA3WT (red), iii) PCNA1WT, PCNA2L171C and PCNA3R112C (blue), and iv) PCNA1G108C, PCNA2WT and PCNA3T180C (green) from a HiLoad 16/600 Superdex 200 pg size exclusion column in 50 mM potassium phosphate buffer, pH 7.4, containing 150 mM KCl, 1 mM EDTA and 10 mM DTT. PCNA3 proteins contained in the mixtures were 1.2 times more than PCNA1 and PCNA2. 14 Figure S3. Elution profiles of the glutathione-treated heterotrimers composed of i) PCNA1WT, PCNA2WT and PCNA3WT (black), ii) PCNA1P186C, PCNA2E105C and PCNA3WT (red), iii) PCNA1WT, PCNA2L171C and PCNA3R112C (blue), and iv) PCNA1G108C, PCNA2WT and PCNA3T180C (green) from a Superdex 200 10/300 GL size exclusion column in 50 mM potassium phosphate buffer, pH 7.4, containing 150 mM KCl. Figure S4. Elution profiles of the mixtures of i) PCNA1WT, PCNA2WT and PCNA3WT (black), ii) PCNA1P186C, PCNA2E105C/L171C and PCNA3R112C (red), iii) PCNA1G108C, PCNA2L171C and PCNA3R112C/T180C (blue), and iv) PCNA1G108C/P186C, PCNA2E105C and PCNA3T180C (green) from a HiLoad 16/600 Superdex 200 pg size exclusion column in 50 mM potassium phosphate buffer, pH 7.4, containing 150 mM KCl, 1 mM EDTA and 10 mM DTT. PCNA prtoeins3s contained in the mixtures were 1.2 times more than PCNA1 and PCNA2. 15 Figure S5. Elution profiles of the glutathione-treated heterotrimers composed of i) PCNA1WT, PCNA2WT and PCNA3WT (black), ii) PCNA1P186C, PCNA2E105C/L171C and PCNA3R112C (red), iii) PCNA1G108C, PCNA2L171C and PCNA3R112C/T180C (blue), and iv) PCNA1G108C/P186C, PCNA2E105C and PCNA3T180C (green) from a Superdex 200 10/300 GL size exclusion column in 50 mM potassium phosphate buffer, pH 7.4, containing 150 mM KCl. a) b) Figure S6. UV-vis specta of a) PCNA1G108C-PdR (solid line), PCNA2L171C-PdX (dashed line) and PCNA3R112C/T180C-P450cam (dotted line), and b) PCNA1WT-PdR (solid line), PCNA2WT-PdX (dashed line) and PCNA3WT-P450cam (dotted line). 16 Figure S7. UV-vis specta of dsPUPPET (solid line) and nPUPPET (dashed line). Supporting References [S1] H. Hirakawa, T. Nagamune, ChemBioChem 2010, 11, 1517-1520. [S2] H. Hirakawa, N. Kamiya, T. Nagamune, Protein Eng. Des. Sel. 2007, 20, 453-459. [S3] I.F. Sevrioukova, C. Garcia, H. Li, B. Bhaskar, T.L. Poulos, J. Mol. Biol. 2003, 333, 377-392. [S4] R. Bhattacharyya, D. Pal, P. Chakrabarti, Protein Eng. Des. Sel. 2004, 17, 795-808. 17