membrane linkage
advertisement

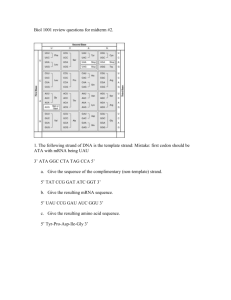
Biology oral exam 2006-07 semifainal 1. Mono-and disaccharides Monosaccharides which typically contain from three to seven carbon atoms are the sugars than cannot be degraded by hydrolysis to simpler sugar. Classify Triose (3 carbons) → glyceraldehyde (C3H6O3; constituent of neutral fat, intermediate of glycolysis) Pentose (5 carbons) → ribose (C5H10O5; constituent of RNA), deoxyribose (C5H10O4; constituent of DNA) Hexose (6 carbons) → glucose and galactose (C6H12O6; they are epimers), fructose (C6H12O6) Aldose; C1=aldehyde → glucose, galactose Ketose; C2=ketone → fructose, dihydroxyacetone Structure Chain form Ring form Aldose CH2OH Aldose (glucose) ketose (fructose) H H C O H C OH H C OH HO C H H C H H O HO C H OH H C OH C OH H C OH C H OH H C H OH O H OH OH OH (α-D-glucose) CH2OH OH (α-D-fructose) CH2OH H+ O C O O OH - H C O OH C OH H OH OH OH OH - OH OH OH O C OH HO2HC O OH CH2OH H CH2OH O C CH2OH ketose H+ OH OH OH OH α-glucose OH β-glucose A disaccharide contains 2 monosaccharides joined by glycosidic linkage. Maltose (molt sugar); α-glucose +α-glucose (α-1,4-glycosidic linkage) Sucrose (table sugar);α-glucose +β-fructose (α-1,2-glycosidic linkage) CH2OH O OH O-H HO OH OH CH2OH O O OH OH OH Condensation Hydrolysis HO2HC (cellobioses make up cellulose) 2. Polysaccharides CH2OH O C4 OH Lactose (milk sugar);β-galactose +β-glucose (β-1,4-glycosidic linkage) OH Cellobiose; β-glucose +β-glucose (β-1,4-glycosidic linkage) C1 O OH OH OH Polysaccharides are the polymer of monosaccharides. Starch → stored energy in plants Amylose: contain onlyα-1,4-glycosidic linkage Amylopectin: containα-1,4-glycosidic linkage and α-1,6-glycosidic linkage (branched) Glycogen → stored energy in animals More branched than starch Cellulose → cell wall of plants Modified saccharides: Galactosamines, N-acetyl glucosamines(chitin), Glycoproteins, Glycolipids OH 3. Lipids ・Hydrophobic organic compunds ・Energy storage ・Component of cellular membrane, bile, hormone 6 main groups ①Neutral fats Fatty acids Glycerol O Usually triacylglycerol (triglyceride) 1 glycerol + fatty acids (usually three) Joined by ester bonds 2 types of fatty acids ・Saturated fatty acids; no double bonds ・Unsaturated fatty acids; contain one or more double bonds ②Phospholipids 2H C O C O H C O C O 2H C O C ester bond Fatty acids Glycerol 1 glycerol + 2 fatty acids + 1 phosphate group Amphipathic O 2H C O C O H C O C C H2 Main component of cell membrane ③Carotenoids P Isoprene hydrophobic hydrophilic β-carotene Pigments (yellow, orange) Consist of isoprene units. Ex. vitamin A (retinal), β-carotene OH O CH2 ④Steroids CH Steran frame Vitamin A Retinal Synthesized from isoprene units Ex. Cholesterol (component of cell membrane), sex hormones, bile, cortisol CH3 ⑤Sphingolipids (CH2)12 Sphingomyelins; sphingosine + phosphate + lecithin Cerebrosides; sphingosine + hexose CH HC CH-OH O CH-NH-C-R →membrane or myelin sheath in nerve tissue CH2-O- - P + lecithin or - glucose ⑥Prostaglandins Unsaturated fatty acid + cyclopentane 4. Amino acids Organic compounds containing amino group and carboxyl group on the same carbon atom. Make up peptides or protein joined by peptide bond. H 20 common amino acids H2N Polar - asparagine(Asn), glutamine(Gln), etc. Acidic - aspartic acid(Asp), glutamic acid(Glu) Contain surfer - cysteine(Cys), methionine(Met) COOH R Non-polar - alanine(Ala), valine(Val), etc. Basic - arginine(Arg), lysine(Lys), histidine(His) C O H2N C C O N C COOH H peptide bond 10 essential amino acids (adult; 9, child; 10) バス雨降り一色 バ(Val; valine)・ス(Thr; threonine)・ア(Arg; arginine→child only)・メ(Met; methionine)・フ(Phe; phenyl alanine)・ リ(Lys; lysine)・ヒ(His; histidine)・ト(Trp; tryptophan)・イ(Ile; isoleucine)・ロ(Leu; leucine) 5. Proteins Basically polymer of amino acids (polypeptide) making up globular or fiber proteins. 4 levels of structure ①Primary Sequence of amino acids joined by peptide bond. ②Secondary Regular (repeating) structure Hydrogen bond between amino acids α-helix; elastic β-pleated sheet; flexible but not elastic triple helix; collagens ③Tertiary 3-D structure of the whole polypeptide The bonds stabilizing tertiary structure of protein are; Ionic bond Disulfide bond (-S-S-) Hydrogen bond Van der Waals force ④Quaternary More than 2 polypeptide chains (subunits). Simple protein; composed of only amino acids Complex protein; not only amino acids. → prosthetic group other organic compounds (saccharides, lipids) Metal Domain → the parts responsible for function. Primary structure determines 2, 3, 4 structures helped by chaperons (heat shock protein). Ex. sickle-cell anemia glutamic acid (ionic)→(change to)→ valine (hydrophobic)→ less soluble in water→ hemoglobin crystallized External factors result in changes Denaturation (irreversible coagulation); heat, heavy metals, strong acid Reversible coagulation; light metals, weak acid 6. DNA Antiparallel, double stranded (double helix) polynucleotide (hetero polymer). Contains genetic information coded in specific sequence of bases. Structure; Each nucleotide composed of 1 base, 1 sugar (pentose), 1 phosphate ・base purine; A, G pyrimidine; T, C ・pentose; deoxyribose ・phosphate purine pyrimidine N N N adenine guanine N O P O N O CH2 N tymine cytosine A base and a pentose together called nucleoside 5’ → nucleotide = nucleoside + monophosphate. Nucleotides are joined by phosphodiester bonds. Complimentary base pairing A = T (2 hydrogen bonds), G ≡ C (3 hydrogen bonds) N N N O N nucleoside O nucleotide Phosphodiester bond O P O O N CH2 O N 3’ O 7. RNA A family of single stranded polynucleotide that function mainly in protein synthesize. Structure; similar to the DNA Differences ・ribose (instead of deoxyribose in DNA) ・Uracil (instead of Tymine in DNA) 3 types of RNA ①m-RNA (messenger RNA); transcribe genetic information from DNA ②t-RNA (transfer RNA); bring amino acids to the ribosome. Have anticodon. ③r-RNA(ribosomal RNA);component of ribosome which catalyze the transformation of amino acids(ribozyme) 8. ATP and coenzymes (1)ATP Energy source of an organism. Contains chemical energy between phosphate; macroenergic bond adenine NH2 N N N ribose O N Structure adenosine H2C O P O O O O O P O P O O O Adenosine (adenine + ribose) + triphosphate ATP is synthesized through the cell respiration. OH OH ATP releases energy as exergonic reaction. ATP → ADP + Pi + Energy This energy is used in endergonic reactions. (2)Coenzyme The organic, detachable cofactors which attach to the allosteric site of apoenzymes. Regulate the activity of enzymes. Ex. NAD+, FAD, Coenzyme A → see also 24. triphosphate 9. Comparison of prokaryotic cell and eukaryotic cell Prokaryotes Eukaryotes YES endomembrnane system NO (nuclear envelope, ER, golgi, lysosome, chloroplast, mitcondria) chromatin with proteins DNA structure circular mitotic spindle NO YES ribosome units 70S (30S + 50S) 80S (40S + 60S) (histone, scafolding protein) 10. The cell nucleus Control center of the cell Prokaryotes; no envelope, floating in cytosol. Eukaryotes; Nuclear envelope (double membrane) Lamina fibrosa (one of intermediate filament) attaching inner envelope stabilizes the structure. Pores; responsible for selective transport of materials (RNA, protein, etc.) Outer envelope is connected to the ER Nucleolus Chromatin structure (DNA+protein) euchromatin (not visible) Hetelochromatin (visible) Nucleoplasm 11. Rough endoplasmic reticulum An organelle participating in endomembrane system. Its cytosolic surface looks like rough because many ribosomes attach to the RER. Responsible for modification of polypeptides. →N-glycosilation (adding saccharides to the nitrogen of side chain of certain amino acids, mostly asparagine) Polypeptide (synthesized by ribosomes using the information of m-RNA) with SRP (signal recognition particle) at its head is injected to the lumen (inner space of RER). →see also 14. 12. Smooth endoplasmic reticulum An organelle participating in endomembrane system. Its surface looks like smooth because ribosomes do not attach. Responsible for lipids synthesis phospholipids fatty acids steroids detoxification →large amount in liver cells. 13. The Golgi complex medial cysteinae An organelle participating in endomembrane system. Responsible for transport of polypeptides; ・vesicles with glycosilated polypeptide from RER fuse with cis-face of golgi. ・modified polypeptides in golgi go to their termination through trans-face of golgi. cis-face trans-face →see also 14. 14. Protein traffic with in the cell Cytosol → RER (rough endoplasmic reticulum) ①Proteins (polypeptides) are produced by ribosome using the information of RNA in cytosol. ②SRP (signal recognition particle) binds to the signal sequence of 5’ end of polypeptide. ③SRP binds to the receptor on RER. ④Polypeptide is injected into lumen of RER through translocon (channel protein). ⑤Signal sequence is removed by enzyme (signal peptidase) ⑥Modification of protein. (N-glycosilation, pruning (removal of some sugars), addition of other sugars) ① Signal sequence Golgi-apparatus lumen Cis-face ② RER SRP (Signal recognition particle) Translocon (channel protein) Receptor for SRP phosphorylation Ribosome core ③ m-RNA polypeptide polypeptide polysaccaride vesicle RER → Golgi-apparatus Polypeptide with polysaccaride is packaged into vesicle. Vesicle fuses with cis-face of Gorgi-apparatus. Lysosome 3 4 Signalize (phosphorilation, pruning, addition of sugars). 2 1 1.default (no-signal addition) → cell membrane 2.M-6-P (Mannose-6-Phosohate) → lysosome 3.special signals → RER (back to the RER) 4.special signals → stay in Gorgi 5.special signals → outside the cell (export protein) Signalized protein reaches their terminal. 15. The lysosomes Lysosomes are organelles that contain digestive enzymes (acid hydrolases). They digest excess or worn out organelles, food particles, and engulfed viruses or bacteria. Lysosomes can fuse with vacuoles and dispense their enzymes into the vacuole, digesting its contents. (Primary lysosome + endosome → socendary lysosome) The interior of the lysosomes (pH 4.8) is more acidic than the cytosol (pH 7). (The constant pH of 4.8 is maintained by proton pumps and Cl- ion channels.) 5 Lysosomal enzymes are synthesized in the rough endoplasmic reticulum (RER) and modified by N-glycosilation. (adding sugars to the Nitrogen of R-chain of certain amino acids (Asparagine)) →Transported and processed through the Golgi apparatus where they receive a mannose-6-phosphate tag (phosphorilation of mannose) that targets them for the lysosome. 16. Mitochondria Mitochondria are the membrane-enclosed organelle, found in most eukaryotic cells. Structure A mitochondrion contains inner and outer membranes. The inner mitochondrial membrane contains proteins with four types of functions; 1. Those that carry out the oxidation reactions of the respiratory chain. 2. ATP synthase, which makes ATP in the matrix. 3. Specific transport proteins that regulate the passage of metabolites into and out of the matrix. 4. Protein import machinery. The inner mitochondrial membrane is compartmentalized into numerous cristae, which expand the surface area of the inner mitochondrial membrane, enhancing its ability to generate ATP. The matrix is the space enclosed by the inner membrane. The matrix contains a highly concentrated mixture of hundreds of enzymes, which the major functions include oxidation of pyruvate and fatty acids, and the citric acid cycle, ribosomes, tRNA, and several copies of the mitochondrial DNA genome. Functions ・Production of ATP (convert organic materials into cellular energy in the form of ATP) This is done by oxidizing the major products of glycolysis: pyruvate and NADH that are produced in the cytosol. This process of cellular respiration, also known as aerobic respiration. ・Apoptosis-programmed cell death ・Heme synthesis ・Steroid synthesis 17. Cytoskeleton Cytoskeleton is a dense network of protein fibers, gives cells mechanical strength, shape, and their ability to move. Also function in cell division and in the transport of materials with in the cell. Cytoskeleton is dynamic internal framework made of 3 types of protein filaments; microtubules, microfilaments, and intermediate filaments. Both microtubules and microfilaments are formed from beadlike, globular protein subunits, which can be rapidly assembled and disassembled. Intermediate filaments are made from fibrous protein subunits and are more stable than the others. (1) Microtubules Hollow cylindrical fibers consisting of tubulin protein subunits (α and β-tubulin), major components of the cytoskeleton and found in mitotic spindles, cilia, flagella, centrioles, and basal bodies. MAPs (microtubule associated proteins) Structural proteins helps regulate microtubule assembly and cross-link microtubules to other cytoskeletal polymers. Motor proteins, kinesin and dynein, produce movement by using ATP. (2) Microfilament Thin fibers consisting of actin protein subunits, are important in cell movement. They are found in muscle cells associated with another protein, myosin. In cytokinesis, they produce cleavage furrow to separate into two daughter cells. (3) Intermediate filament They are intermediate in size between microtubules and microfilaments. They strengthen the cytoskeleton and stabilize cell shape, examples are lamina fibrosa inside the nuclear envelope, neuron filaments in axons of neuron cells, and keratin in hairs, nails, and skin. 18. Centrosome In animal cells, centrosome is the main part of MTOCs (microtubule-organizing centers), the region of the cell from which microtubules are anchored and possibly assembled. It contains 2 centrioles which are oriented at right angles to each other. (each centriole has a 9×3 structure consisting of 9 sets of three attached microtubules arranged to form a hollow cylinder) During the cell division, centrosomes move to the poles of the cell and make up the mitotic spindle. 19. The cell membrane The cell membrane is composed of phospholipid bilayer, cholesterols, proteins, and carbohydrates. ①Phospholipids (lipids in which 2 fatty acids and a phosphorous- containing group are attached to glycerol) They associate as bilayers in water because they are roughly cylindrical amphipathic molecules (The hydrophobic fatty acid chains associate with each other and are not exposed to water. The hydrophilic phospholipid heads are in contact with water.) The ordered arrangement of phospholipid molecules makes the cell membrane a liquid crystal. (The hydrocarbon chains are in constant motion, allowing each molecule to move laterally on the same side of the bilayer.) Various transport and secretary vesicles form from phospholipid bilayers and also merge with membranes of the ER and Gorgi complex, facilitating the transfer of materials from one compartment to another. Phospholipid bilayer presents a barrier to most polar molecules because the interior of it is hydrophobic. ②Cholesterol Cholesterols play a role as a fluidity buffer. At low temperature, they act as “spacers” between the hydrocarbon chains, restricting van der Waals interaction that would promote solidifying. At high temperature, they connect hydrophilic parts and stabilize membrane. ③Proteins The 2 major classes of membrane proteins are Integral proteins and Peripheral proteins. (1)Integral proteins They are firmly bound to the membrane. They are amphipathic. (their hydrophilic regions extend out of the cell or into the cytoplasm, while their hydrophobic regions interact with the fatty acid tails of the membrane phospholipids (α-helix). Transmembrane proteins; extend completely through the membrane. Non-transmembrane proteins; do not extend all the way through the membrane. (2)Peripheral proteins They are located on the inner or outer surface of the membrane, usually bound to exposed regions of integral proteins by noncovalent interactions. Functions of membrane proteins are; Anchoring (integrin), transport (channel protein, carrier protein), enzymatic activity (membrane-bound enzyme), signal transduction (receptor), cell recognition (antigen), intercellular junction. ④Hydrocarbons They are exposed on the extracellular surface, and play roles in cell recognition and adhesion as glycolipids (carbonhydrates attached to lipids) or gycoproteins (carbonhydrates attached to proteins). 20. Membrane transport (1)Passive transport; does not require energy ①Diffusion (or simple diffusion) The net movement of particles (atoms, molecules, or ions) down its concentration gradient from a region of greater concentration to one of lower concentration. (gases (O2, CO2, N2), small polar molecules (H2O, glycerol), larger nonpolar substances (hydrocarbons) Slightly larger polar molecules (glucose) and charged ions of any size are also pass through but slowly.) ②Facilitated diffusion The passive transport of ions or molecules by a transport protein in membrane (channel protein, carrier protein) As in simple diffusion, net transport is down a concentration gradient, and no additional energy has to be supplied. (ions, amino acids, sugars, water-soluble molecules) (2)Active transport; require energy ①Primary active transport (carrier mediated active transport) The transport of ions or molecules across a membrane against a concentration gradient or electrical gradient. It requires both a transport protein with binding site for the specific substance and energy directly supplied by ATP. (Na+-K+ pump, H+ pump (lysosome)) ②Secondary active transport (cotransport) The active transport of one substance against a concentration gradient by coupling its transport to the transport of another down its concentration gradient. It requires energy but not from ATP directly. (symport; Na+ and glucose in the intestine antiport; Na+ and Ca2+) 21. Types of endocytosis High concentration Na+ Low concentration glucose outside inside High concentration High concentration Na+ Ca2+ Na+ Ca2+ outside Na+ Low concentration inside glucose High concentration Low concentration Low concentration Endocytosis is the active transport of large substances into the cell by the formation of cytoplasmic vesicles or vacuoles enclosing the material, and then the material is released inside the cell. There are 3 types of endocytosis; phagocytosis, pinocytosis, and receptor-mediated endocytosis. ①Phagocytosis (literally cell eating) The plasma membrane encloses a large solid particles, such as a bacterium or food, forms a vacuoles around it, and moves it into the cell. The vacuoles then fuses with lysosomes, and ingested material is degraded. ②Pinocytosis (literally cell drinking) The cell takes in dissolved materials. Tiny droplets of fluid are trapped by folds in the plasma membrane, which pinch off into the cytosol as tiny vesicles. The liquid contents of these vesicles are then slowly transferred into the cytosol. ③Receptor-mediated endocytosis →see 22 22. Receptor-mediated endocytosis A type of endocytosis in which extracellular molecules become bound to specific receptors on the cell surface, and then enter the cytoplasm enclosed in vesicles. ①Ligand (a molecule that binds specifically to a receptor) binds to receptors in coated pits of membrane. ②Coated vesicle forms by endocytosis. ③Coating detaches from vesicle. ④Contents are transferred to endosome. ⑤Ligand separates from its receptors. ⑥Endosome fuses with lysosome. ⑦Contents are digested and released into cytosol. ※Receptors are transported to membrane and recycled Cells take up cholesterols from the blood by this process. ② Ligand (LDL particle) Clathrin recycled ① ③ LDL receptor Clathrin ④ ⑤ Endosome Lysosome Free cholesterol ⑦ ⑥ ※receptor recycled 23. Endergonic and exergonic reactions Exergonic reactions (spontaneous; energy releasing) An exergonic reaction releases energy. Free energy reactants energy The total free energy in its final state is less than the products total free energy in its initial state. [⊿G is negative] Ex) catabolisum Endergonic reaction (not spontaneous; energy requiring) An endergonic reaction is a reaction in which there is gain of the free energy. Free energy energy products The free energy of the products greater than the free energy of the reactant. Ex) anabolisum [⊿G is positive] reactants 24. Enzymes (structure, role) Biological catalysts active site Increase the speed of the chemical reaction substrate apoenzyme without being consumed by the reaction. Decrease the activation energy. consist of only protein. Ex. pepsin have two component; apoenzyme + cofactor cofactor allosteric site Neither the apoenzyme nor cofactor alone has catalytic activity. ※Cofactor; additional chemical component inorganic; metals. Ex.magnesium ion, calcium ion, iron, copper, zinc organic coenzyme. Ex. NADPH, NADH, FADH2 ATP prosthetic group. Ex. heme ※Coenzyme: An organic, nonploypeptide compound that binds to the apoenzyme and serves as a cofactor 25. Regulation of enzymatic activity Activation of proenzyme. Ex. pepsinogen → pepsin Allosteric modulation. Ex. cAMP dependent protein kinase (cAMP removes allosteric inhibitor) feed back inhibition Phosphorylation Genetic control induction repression Inhibition of enzyme Reversible inhibition competitive inhibition noncompetitive inhibition →(some feature in common with allosteric inhibition) Irreversible inhibition Feedback inhibition 1 Reversible inhibition (competitive, noncompetitive inhibition) ●competitive inhibition In competitive inhibition, the inhibitor competes with the normal substrate for binding to the active site of the enzyme ●noncompetitive inhibition In noncompetitive inhibition, the inhibitor binds with the enzyme at site other than active site and noncompetitive inhibition has some feature in common with allosteric inhibition 2 Irreversible inhibition In irreversible inhibition, an inhibitor permanently inactivates or destroys an enzyme. (Ex)inhibitor …heavy metals (Hg, Pb ) 3 Feedback inhibition A type of enzyme regulation in which the accumulation of the product of a reaction inhibits an earlier reaction in the sequence also known as product inhibition. 26. Steps of aerobic respiration The chemical reaction of the aerobic respiration of glucose are grouped into four stage that are Glycolysis, Formation of acetyl CoA, Citric acid cycle and Electron transport chain. In eukaryotes, the first stage take place in the cytosol and the rest stages take place inside mitochondria. During aerobic respiration, nutrients are catabolized to carbon dioxide and water. ①Glycolysis Series of reaction in which glucose is degraded to pyruvate. (net profit of 2ATP;hydrogen atoms are transferred to carries; can proceed anaerobically ) ②Formation of acetyl CoA Pyruvate is degraded and combined with coenzyme A to form acetyl CoA (hydrogen atoms are transferred to carries; CO2 released) ③Citric acid cycle Series of reactions in which the acetyl portion of acetyl CoA is degraded to CO2 (hydrogen atoms are transferred to carries; ATP is synthesized) ④Electron transport chain Chain of several electron transport molecules; electron are passed along chain; released energy is used to form proton gradient; ATP synthesized as proton diffuse down gradient; oxygen is final electron acceptor. glucose 2ATP ④Electron transport chain G-3-P ×2 ①Glycolysis 4ATP 2NAD+ 2NADH+H+ H+ + pyruvate ×2 cytosol 1 O + e2 2 H2O mitochondria ②Formation of acetyl CoA ADP + Pi 2CO2 2coA 2NAD+ ATP 2NADH+H+ acetyl-coA ×2 citrate (C6) oxaloacetate (C4) ×2 ③Citric acid cycle 2ATP 6NADH+H+ 2FADH2 27. Glycolysis Glycolysis is the first stage of respiration, literally splitting sugar. In glycolysis, glucose is converted to two molecules of pyruvate in the cytosol and there is a net yield of 2 ATP and 2 NADH molecules Glycolysis is divided into two major phases, the energy investment phase and the energy capture phase. Glycolysis C6H12O6 + 2 NAD++ 2ADP → 2C3H4O3 + 2NADH + 2 ATP Glucose pyruvate (1)The energy investment phase (=endergonic reaction ) ①In two separate phosphorylation reaction, a phosphate group is transferred from ATP to the sugar. →substrate-level phoshorylation; ATP formed when a phosphate group is transferred to ADP from phosphorylated intermediate. [glucose → fulctose-1,6-bisphosphate] 2 ATP are used ②The fulctose-1,6-bisphosphate is converted to 2 glyceraldehyde phosphate(G3P) (2)The energy capture phase (exergonic reaction) G3P is converted to pyruvate. 4 ATP and 2 NADH + H+ are produced. NADH + H+ produced in glycolysis is used in electron transport chain. Cells in the heart, the liver, the kidney Electrons stored in NADH + H+ are transported through malate-asparatate shuttle. → 3 ATP are synthesized from 1 NADH + H+ Cells in others Electrons stored in NADH + H+ are transported through glycerophosphate shuttle. → 2 ATP are synthesized from 1 NADH + H+ 28. Citric acid cycle Citric acid cycle is series of chemical reaction in aerobic respiration. In citric acid cycle, acetyl coenzyme A is completely degraded to carbon dioxide and water (NADH, FADH2) with the release of metabolic energy that is used to produce ATP. To summarize, citric acid cycle yields 4CO2, 6 NADH, 2 FADH2 and 2 ATP per glucose molecule. Citric acid cycle Acetyl CoA + 6 H2O → 4 CO2 + 6 NADH + 2 FADH2 + 2 ATP electron transport chain NADH+H+ acetyl-coA C2 citrate C6 C4 isocitrate oxaloacetate FADH2 C4 NADH+H+ C6 C4 malate C4 fumarate succinate 8 H2O ADP ATP GTP GDP α-ketoglutarate C5 succinyl-coA C4 NADH+H+ 29. The electron transport chain The series of redox reactions which hydrogens or their electrons are passed along from one electron acceptor to another, finally electrons are accepted by oxygen and form water. During these reactions, protons are transported actively from matrix to inter membrane space, causing a concentration gradient of H+. H+ Concentration gradient causes energy for ATP synthesis by passing through the enzyme complex. (ATP synthase) NADH + H+ donates its electrons to the complexⅠ → 3 ATP are produced from 1 NADH + H+ FADH2 donates its electrons to the complexⅡ → 2 ATP are produced from 1 FADH2 H+ H+ H+ intermembrane space H+ H+ H+ H+ H+ H+ complexⅠ CoQ NAD+ H+ Ⅲ Ⅳ H+ 2H+ complexⅡ H+ NADH+H+ FADH2 FAD matrix H+ H+ H+ 1 O 2 2 H+ H+ H+ inner membrane H2O ATP synthase H+ ADP + Pi ATP 30. Fermentation An anaerobic process for producing ATP Final electron acceptor is organic compound, not oxygen. ①Lactate fermentation; microbes, human(muscle cells, red blood cells) glucose cytosol 2ATP G-3-P ×2 4ATP 2NADH+H+ 2NAD+ 2NAD+ pyruvate ×2 lactate ×2 Lactates are formed from pyruvates in order to oxidize NADH + H+ to NAD+, and NAD+ are recycled. ②Alcohol fermentation; microbes(bacteria, fungi) glucose cytosol 2ATP G-3-P ×2 4ATP 2NAD+ 2NADH+H+ 2NAD+ ethanol ×2 pyruvate ×2 CO2 Ethanols are formed from pyruvate in order to oxidize NADH + H+ to NAD+, and NAD+ are recycled. 31. The catabolic pathways of proteins and fats in the cell Protein consist of amino acid and that are metabolized by reaction in which the amino group (-NH2) is first removed, a process called deamination. Therefore amino acid divided into amino group and carbon chain. The amino group is converted to urea and excreted, but the carbon chain is used as a reactant in one of step of aerobic respiration such as Glycolysis, Acetyl CoA and Citric acid cycle. Ex) The catabolic pathway of proteins Carbon chain Step of aerobic respiration Glycolysis alanine ⇒ glutamate ⇒ Citric acid cycle aspartate ⇒ Citric acid cycle leucine ⇒ acetyl CoA Fat which consists of Glycerol and fatty acid are also used fuel. Phosphate is add to Glycerol, converting it to G3P (glyceraldhyde-3-phosphate) or another compound that enters glycolysis. Fatty acids are oxidized and split enzymatically into two-carbon acetyl groups that are bond to coenzyme A. This process which occurs in the mitochondrial matrix is called β-oxidation. 32. Comparison of anabolic and catabolic reactions in the cell Metabolism Anabolism; simple → complex (endergonic) Primary; inorganic → organic (photosynthesis; plants only) Secondary; organic → organic (glucose → starch, glycogen; plants, animal, fungi) Catabolism; complex → simple (exergonic) Cellular respiration 33. The cell cycle The cell cycle is the period from the beginning of one division to the beginning of the next. Interphase(G1 phase → S phase → G2 phase) → M phase ①G1 phase (first gap phase) The cell is carrying out its normal function. G1 Euchromatin state. (not visible) S 46 single chromosomes → 2n (23 paternal and 23 maternal) M G2 46 DNA ②S phase (synthesis phase) DNA is replicated. M phase (Mitosis and cytokinesis) Still euchromatin state. 46 double chromosomes → 2n (23 paternal and 23 maternal) 92 DNA ③G2 phase (second gap phase; shorter than G1 phase) Enzymes for mitosis are created. Single chromosomes G1 S G2 2n 2n 2n M 2n Still euchromatin state. 2n 46 double chromosomes → 2n (23 paternal and 23 maternal) Double chromosomes 92 DNA ④M phase (mitosis phase; contains cytokinesis) Prophase → metaphase → anaphase → telophase → cytokinesis → see 40. Cells are diploid cells (homologue chromosomes; 2 sets of chromosomes (1 paternal and 1 maternal)) through the cell cycle. Cell cycle is controlled by cyclins (regulatory proteins) and cyclin-dependent kinase (protein kinase) 34. DNA synthesis (= replication) The process by which DNA is duplicated. Origin of replication Semiconservative. ⑥DNA polymeraseⅢ Enzymes needed for replication are; ①topoisomerase → nicks in DNA (relining tension in DNA) ②DNA helicase → unwound DNA strands. ③SSB (single strand binding protein) or helix destabilizes protein ①topoisomerase → single strand DNA is fixed by this enzyme RNA primer ④DNA primase → synthesize short RNA primer ③SSB ⑤DNA polymeraseⅠ → gap filling in the places between okazaki fragments. ②DNA helicase ⑥DNA polymeraseⅢ → synthesize new DNA strand by adding nucleoside triphosphate (or nucleotide diphosphate) (DNA polymeraseⅡ is needed for repairing of DNA) ⑦DNA ligase → connect DNA strand and the short section replaced Direction of replication by DNA polymeraseⅠ 5’ 5’ 3’ A 3’ A T C C A T OH Leading strand G G C T A 3’ 5’ OH 3’ 5’ T C G G 3’ (bidirectional) 5’ 3’ Lagging strand (Okazaki fragments) 3’ 5’ 5’ 3’ 3’ 5’ RNA primer 5’ 5’ 5’ ①RNA primer is made by DNA primase Direction of replication ②new strands are synthesized by DNA polymeraseⅢ 3’ 5’ 5’ 5’ space of it is replaced by DNA polymeraseⅠ 3’ 3’ ③RNA primer is degraded, ④strands are combined by DNA ligase 5’ 3’ 5’ 3’ 5’ 3’ Telomeres; end part of DNA (repeating, non-transcribed sequences) At each replication, telomeres get a bit shorter. First primer is not replaced by DNA DNA polymeraseⅠ → telomeres are used up after a certain number of replications → cells die after a limited number of cell divisions 3’ 5’ 5’ 3’ ligase 3’ 5’ 5’ 3’ End of DNA 3’ 5’ 5’ 3’ telomere 3’ 5’ 5’ 3’ 35. Transcription in prokaryotes Transcription is the synthesis of RNA complementary to the template DNA strand. m-RNA contains information that specifies the amino acid sequences of polypeptide chains. Transcription in prokaryotes occurs in the cytoplasm alongside translation. ①Initiation ・RNA polymerase (RNAP) recognizes and specifically binds to the promoter region on DNA. (promoter is not transcribed) At this stage, the DNA is double-stranded ("closed"). This RNAP/wound-DNA structure is referred to as the closed complex. ・The DNA is unwound and becomes single-stranded ("open") in the Template strand of DNA promoter ②Elongation 5’ 3’ Translated region mRNA 3’ 5’ vicinity of the initiation site. This RNAP/unwound-DNA structure is called the open complex. mRNA termination Transcribed sequence region leader sequences ・mRNA is elongated until RNAP recognizes a stop signal termination trailing sequences Start codon Stop codon sequence. ・New nucleotide is added to the 3’ end of growing chain by RNAP similar polypeptide to the DNA polymerase in DNA replication. ③Termination ・Transcription stops at the end of the stop signal. When RNAP comes to the stop signal, it releases both the DNA template and the new RNA strand. (in eukaryotic cells, RNAP adds nucleotides to the mRNA after it passes the stop signal) Transcription does not require primer, but needs several proteins to initiate. In prokaryotes, transcription and translation are coupled. →ribosomes bind to the 5’ end of the growing mRNA and initiate translation before the mRNA is fully transcribed. Usually degradation of the 5’ end of mRNA begins even before the polypeptide is complete. The half-life (the time it takes for half the molecules to be degraded) of mRNA in prokaryotes is only about 2 minutes. 36. Transcription in eukaryotes In prokaryotes, mRNA is translated as it is transcribed from DNA. However, this cannot occur in eukaryotes because eukaryotic chromosomes are confined to the cell nucleus, and protein synthesis take place in the cytosol. Before it is translated, eukaryotic mRNA is transported through the pores in the nuclear envelope and into the cytosol. Before mRNA is transported from nucleus, posttranscription occurs. (→see under) 5’ cap and poly-A tail are considered that they help export the mRNA from the nucleus, stabilize against degradation in the cytosol, and facilitate initiation of translation. Eukaryotic mRNAs are much more stable than prokaryotic one. Their half-lives range from 30 minutes to as long as 24 hours. The average half-life of a mammalian cell is about 10 hours. Pre-mRNA Start codon ①A DNA sequence containing both exons and introns Stop codon 5’ is transcribed by RNA polymerase to make 3’ G pre-mRNA. exon ②Pre-mRNA is capped by the addition of a modified base intron 5’ 3’ G A (7-metylguanosine cap) to its 5’ end. A ③A poly-A (adenine) tail is added to the 3’ end. mature-mRNA 3’ 5’ G A A ④Splicing (removal of introns and exons are spliced nucleus together). cytosol ⑤The mature mRNA is transported through the nuclear 3’ 5’ G A envelope. A 37. Translation The conversion of information provided by mRNA into a specific sequence of amino acids in a polypeptide chain. This process requires tRNA and ribosomes. ①Initiation Initiation factors bind to the small ribosomal subunit. The small ribosomal subunit with initiation factors binds to mRNA Small rebosomal subunit in the region of start codon (AUG). Start codon 3’ 5’ The initiator tRNA binds to the start codon, and one of the initiation factors is released. The large ribosomal subunit binds to them, remaining initiation mRNA factors are released, the initiation complex is complete. Initiation factor MET (fMET in bacteria) Initiator tRNA Large ribosomal subunit P site A site E site 5’ 3’ 3’ 5’ Initiation complex ②Elongation Cyclic process in which amino acids are added one by one to the growing polypeptide chain. Proceeds in the 5’ → 3’ direction along the mRNA. Polypeptide chain grows from its amino end to its carboxyl end. 3 sites on the ribosome. A site → tRNA with new amino acid P site → tRNA with polypeptide E site → site for exit ①The polypeptide chain is covalently bonded to the tRNA that Amino acids AminoacyltRNA Carries the amino acid most recently added to the chain. This tRNA is in the P site of the ribosome. P E A 5’ 3’ Aminoacyl tRNA binds to codon in A site GTP → GDP Amino acids ②An aminoacyl tRNA binds to the A site by complementary basepairing between the tRNA’s anticodon and the mRNA’s codon. E P A 5’ 3’ Peptide bond formation ③The growing polypeptide chain detaches from the tRNA in the P site, and becomes attached by a peptide bond to the amino Acid linked to the tRNA at the A site. E P A 5’ 3’ Translocation toward 3’ end of mRNA GTP → GDP ④In the translocation step, the ribosome moves one codon toward the 3’ end of mRNA. As a result, the growing polypeptide chain is transferred to the E P P site. A 5’ 3’ Uncharged tRNA in the E site exits the ribosome. Back to ①, and these cycles are continued. ③Termination Termination occurs when the ribosome reaches one of the three stop codons (UAA, UAG, UGA). The A site binds to a release factor. Release factor triggers the release of the completed polypeptide chain and dissociation of the translation complex. Release factor ①When the ribosome reaches a stop codon, the A site binds to a protein release factor. E P A 5’ 3’ Stop codon (UAA, UAG, UGA) Polypeptide chain is released ②The release factor hydrolyzes the bond between the polypeptide chain and the tRNA. A E It causes the release of the polypeptide chain from the tRNA P in the P site. 5’ 3’ Stop codon (UAA, UAG, UGA) ③The remaining parts of the translation complex dissociate E P A 5’ H O H2N C C O R 3’ 3’ 5’ Loop 2 ※tRNA Connection between the mRNA and polypeptide by having Loop 3; aminoacyla triplet codon (anti codon). tRNA 3 loops loop1 → anti codon (complementary to the codon in mRNA) synthetase loop2 → for attachment to the ribosome Loop 1; anticodon loop3 → for aminoacyl-tRNA synthetase (use 2 ATP (ATP → AMP + 2Pi)) 38. Comparison of eukaryotic and prokaryotic cell functions → see 9 → compare 35 and 36 There are 3 types of RNA polymerase in eukaryotes, but only 1 type of RNA polymerase in prokaryotes. RNA polymeraseⅠ; rRNA in big subunit of the ribosome RNA polymeraseⅡ; mRNA RNA polymeraseⅢ; tRNA, and rRNA in small subunit of the ribosome 39. Mutations Types of mutations range from disruption of a chromosome’s structure to a change in only a single pair of nucleotide bases. Mutations that affect the base sequence of the DNA are; (1)Base substitution → 1 base is changed to another (A ⇔ G (purine), C ⇔ T (pyrimidine)) ①Silent mutation 1 base is changed, but the codon codes the same amino acid. Therefore no change in the amino acid sequence and final product (protein). Ex. TCC (the codon in mRNA is AGG which codes Arginine) → (C is changed to T) → TCT (the codon in mRNA is AGA which still codes Arginine) ②Missense mutation 1 base is changed, and the codon codes the wrong amino acid. Ex. sickle cell anemia (the codon coding Glutamic acid (ionic) is changed to another codon coding Valine (hydrophobic), and causes a change in the shape of hemoglobin) ③Nonsense mutation 1 base is changed, and the stop codon is created. Ex. GCT (the codon in mRNA is CGA which codes Arginine) → (G is changed to A) → ACT (the codon in mRNA is UGA which is the stop codon) (2)Frameshift mutation ①Insertion 1 or 2 nucleotides are inserted into the DNA, altering the reading frame. As a result, a new sequence of amino acids is created downstream of the insertion site. ②Deletion 1 or 2 nucleotides are deleted from the DNA, also altering the reading frame. 40. Mitosis The division of cell nucleus resulting in 2 daughter nuclei, each with the same number of chromosomes as the parent nucleus. Cytokinesis usually overlaps the telophase. 4 phases ①Prophase Early prophase ・nuclear envelope begins to disappear. ・nucleolus disappears. ・long fibers of chromatin become evident and begin to condense as visible chromosomes. (each chromosome is duplicated in S phase, composed of a pair of sister chromatids) Late prophase ・chromosomes continue to shorten and thicken. centromere Kinetochore (protein) 2 chromatids (a pair of sister chromatids) ・mitotic spindle begins to form. ②Metaphase ・chromosomes aligned on the metaphase plate. (equatorial plane) ・mitotic spindle is complete 3 types of microtubules in mitotic spindle 1 ①kinetochore microtubules → attach to kinetochore on the chromosomes, and to centrosome (in animal cells, each centrosome contains 2 2 centrioles). 3 Centrosome is also called MTOC (microtubule-organizing center). ②astral microtubules → anchor the centrosomes to the ③pole to pole microtubules → fixation of the mitotic spindle poles. ③Anaphase ・Anaphase begins as the sister chromatids separate. ・The new separate chromosomes move to opposite poles, using the spindle microtubules as tracks. (once the chromatids are separated, each chromatid is called chromosome) ・ Anaphase ends when all the chromosomes have reached the poles. ④Telophase ・Anaphase begins as the sister chromatids separate. ・The new separate chromosomes move to opposite poles, using the spindle microtubules as tracks. (once the chromatids are separated, each chromatid is called chromosome) ・ Anaphase ends when all the chromosomes have reached the poles. Cleavage furrow 41. Meiosis 42. Comparison of mitosis and meiosis 43. Lactose operon model An operon is a group of key nucleotide sequences including an operator, a common promoter, and one or more structural genes that are controlled as a unit to produce messenger RNA (mRNA). Operons occur primarily in prokaryotes and nematodes(線虫). Control of operon genes is a type of gene regulation that enables organisms to regulate the expression of various genes depending on environmental conditions. Operon regulation can be either negative or positive. Negative regulation involves the binding of a repressor to the operator to prevent transcription. Repressor gene Operon Promoter Operator ・Repressor protein is translated and binds to the operator in normal condition. (glucose (+) , lactose(-)) Structural ・When the allolactose (isomer of lactose) presents in the nucleus, Operon Repressor gene Structural gene Promoter Operator DNA Z Y A allolactose binds to the allosteric site of repressor protein. ・Thus, prepressor protein detaches from operator. Allolactose RNA-polymerase can bind to promoter. Promoter Operator DNA ・After detaching of repressor from operator, Operon Repressor gene Z Structural gene Y A transcription RNA- polymerase translation Z Y A ・The structural genes「Z」 「Y」 「A」on DNA are now transcribed and translated.