Exercises Phylogenetic Trees
advertisement
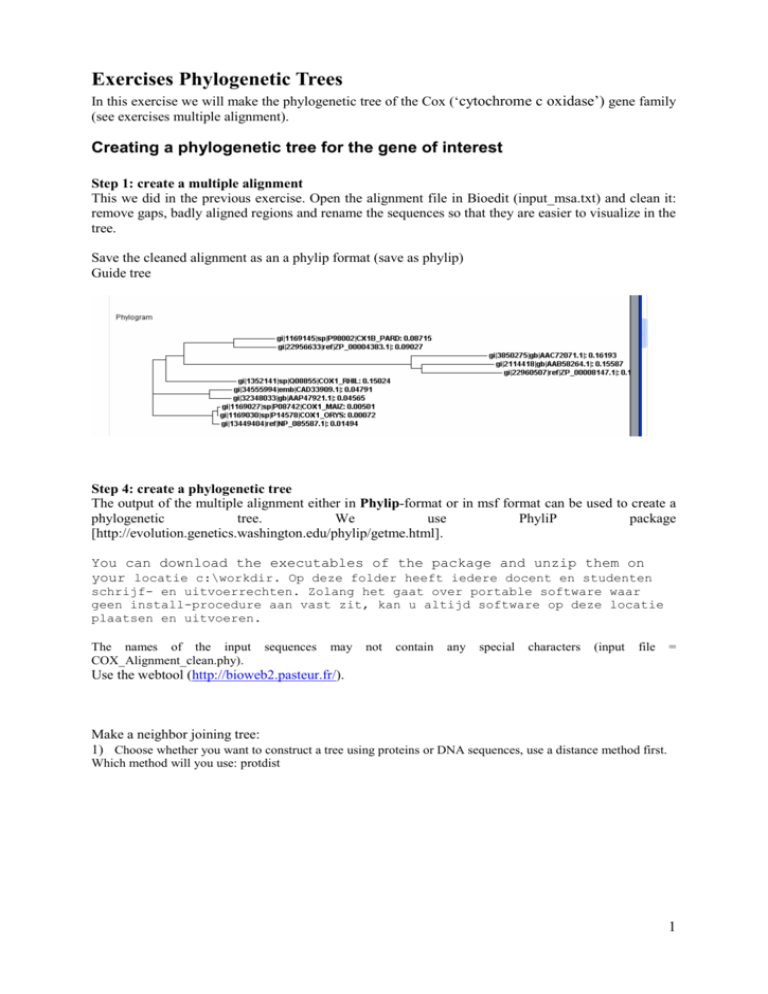
Exercises Phylogenetic Trees In this exercise we will make the phylogenetic tree of the Cox (‘cytochrome c oxidase’) gene family (see exercises multiple alignment). Creating a phylogenetic tree for the gene of interest Step 1: create a multiple alignment This we did in the previous exercise. Open the alignment file in Bioedit (input_msa.txt) and clean it: remove gaps, badly aligned regions and rename the sequences so that they are easier to visualize in the tree. Save the cleaned alignment as an a phylip format (save as phylip) Guide tree Step 4: create a phylogenetic tree The output of the multiple alignment either in Phylip-format or in msf format can be used to create a phylogenetic tree. We use PhyliP package [http://evolution.genetics.washington.edu/phylip/getme.html]. You can download the executables of the package and unzip them on your locatie c:\workdir. Op deze folder heeft iedere docent en studenten schrijf- en uitvoerrechten. Zolang het gaat over portable software waar geen install-procedure aan vast zit, kan u altijd software op deze locatie plaatsen en uitvoeren. The names of the input COX_Alignment_clean.phy). sequences may not contain any special characters (input file = Use the webtool (http://bioweb2.pasteur.fr/). Make a neighbor joining tree: 1) Choose whether you want to construct a tree using proteins or DNA sequences, use a distance method first. Which method will you use: protdist 1 2) paste the input sequences (in phylip format) Protdist outfile 11 CX1B_PARD 0.654107 Cox_RSpae 0.649160 COX1_MAIZ 0.357418 COX1_ORYS 0.357594 COX_ATHA 0.358977 Cox_MMUS 0.084182 COX_HSAP 0.000000 0.000000 0.429304 0.154261 0.460986 0.500779 0.340058 0.501671 0.340134 0.498997 0.345956 0.634077 0.402452 0.654107 0.393297 0.154261 3.134231 0.000000 3.073867 0.485995 3.379677 0.486848 3.379781 0.484239 3.410483 0.618292 3.888354 0.649160 3.892178 0.500779 3.210408 0.485995 3.115685 0.000000 3.306331 0.002319 3.304729 0.016392 3.343217 0.362214 3.819274 0.357418 3.793549 0.501671 3.139896 0.486848 3.169265 0.002319 3.278662 0.000000 3.277049 0.014039 3.308833 0.362392 3.667533 0.357594 3.637363 0.498997 0.634077 0.484239 0.618292 0.016392 0.362214 0.014039 0.362392 0.000000 0.364467 0.364467 0.000000 0.358977 0.084182 2 COX1_RHIL 0.393297 FIXN_RLEG 3.892178 COX_RSPA 3.793549 CYTN_ABRA 3.637363 0.429304 0.000000 3.134231 3.366747 3.210408 3.432779 3.139896 3.452019 0.460986 3.366747 3.073867 0.000000 3.115685 0.290228 3.169265 0.433081 0.340058 3.432779 3.379677 0.290228 3.306331 0.000000 3.278662 0.417514 0.340134 3.452019 3.379781 0.433081 3.304729 0.417514 3.277049 0.000000 0.345956 0.402452 3.410483 3.888354 3.343217 3.819274 3.308833 3.667533 3) Use neighbour joining to cluster the sequences in a tree: (look at the sample file how the input should be, it should be the output of the previous distance calculation (protdist(1).outfile)) 4) view the treefile (use drawtree or the java application archeopterix) download http://www.phylosoft.org/archaeopteryx/ forester.jar double click to start the application Open the outtree file Distance measure and neighbor joining: (Cox_RSpae:0.07320,(((FIXN_RLEG:0.15705,COX_RSPA:0.13317):0.09847, CYTN_ABRA:0.18171):2.81701,(COX1_RHIL:0.15627,((Cox_MMUS:0.04166, COX_HSAP:0.04253):0.22401,((COX1_MAIZ:0.00173,COX1_ORYS:0.00059):0.00336, COX_ATHA:0.01069):0.08783):0.06240):0.20722):0.03727,CX1B_PARD:0.08106); 3 Now make a maximum parsimony tree 4 Maximal parsimony ((((CYTN_ABRA,(COX_RSPA,FIXN_RLEG)),(COX1_RHIL,((COX_HSAP,Cox_MMUS), ((COX_ATHA,COX1_ORYS),COX1_MAIZ)))),Cox_RSpae),CX1B_PARD); [geeft geen taklengtes] View the tree using http://bioinformatics.psb.ugent.be/hypergeny/ Neighbor joining +Cox_RSpae ! ! +--FIXN_RLEG ! +-1 ! +-----------------------------------------2 +-COX_RSPA ! ! ! ! ! +--CYTN_ABRA 4-5 ! ! +-COX1_RHIL ! ! ! 5 ! +--6 +Cox_MMUS ! ! +---3 ! ! ! +COX_HSAP ! +-7 ! ! +COX1_MAIZ ! ! +-8 ! +-9 +COX1_ORYS ! ! ! +COX_ATHA ! +CX1B_PARD Maximal parsimony +----CYTN_ABRA +---------------10 ! ! +-COX_RSPA ! +-9 +-8 +-FIXN_RLEG ! ! ! ! +-------------COX1_RHIL ! ! ! ! +-------7 +-COX_HSAP ! ! +-------6 ! ! ! +-Cox_MMUS +-2 +-5 ! ! ! +-COX_ATHA ! ! ! +-4 ! ! +----3 +-COX1_ORYS 1 ! ! ! ! +----COX1_MAIZ ! ! ! +-------------------------Cox_RSpae ! +----------------------------CX1B_PARD remember: this is an unrooted tree! 6 Make a neighbor joining tree with bootstrapping: First generate 100 random datasets using BootSeq.exe 7 Generate a neighbor joining tree of all the datasets. Make sure you change in the input: the use of multiple datasets Each time you run a different script rename the files so that they do not longer have the default name After running protdist en neighbor joining you will end up with a tree file containing 100 sequences in Newick format. These have to be converted in a consensus tree using the majority rule with the program consensus (note this program works on the outtree output). View the result and interpret the outcome Extended majority rule consensus tree CONSENSUS TREE: the numbers on the branches indicate the number of times the partition of the species into the two sets which are separated by that branch occurred among the trees, out of 100 trees +-------------CYTN ABRA +---------------10.0-| | | +------COX RSPA | +--7.0-| | +------FIXN RLEG +--7.0-| | | +---------------------------COX1 RHIL | | | | | | +-------------COX ATHA | +-10.0-| +-10.0-| | | | | +------COX1 MAIZ +------| | | +--8.0-| 8 | | +--6.0-| +------COX1 ORYS | | | | | | +------Cox MMUS | | +--------10.0-| | | +------COX HSAP | | | +-----------------------------------------CX1B PARD | +------------------------------------------------Cox RSpae CONSENSUS TREE: the numbers on the branches indicate the number of times the partition of the species into the two sets which are separated by that branch occurred among the trees, out of 100.00 trees +-------COX RSPA +--82.0-| +-100.0-| +-------FIXN RLEG | | +--99.0-| +---------------CYTN ABRA | | | | +-------CX1B PARD | +----------61.0-| | +-------Cox RSpae +-------| | | +-------COX1 ORYS | | +--76.0-| | | +--99.0-| +-------COX1 MAIZ | | | | | +--57.0-| +---------------COX ATHA | | | | +-------Cox MMUS | +---------100.0-| | +-------COX HSAP | +---------------------------------------COX1 RHIL 9 After bootstrapping we have to calculate a consensus tree and then the branch lengths are lost. More information on the Phylip package http://evolution.genetics.washington.edu/phylip.html 10