Zoo/Bot 3333
advertisement

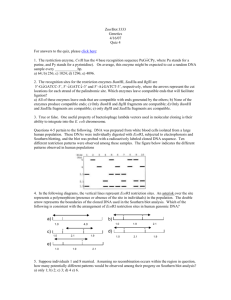
Zoo/Bot 3333 Genetics 4/21/11 Quiz 4 For the answers to the quiz, click here: Questions 1-2 pertain to the following. The ability to find and access information is critical to both scholarship and professional development, and the first two questions below will require you to go to ‘extramural’ sources to find answers to questions relevant to topics we have recently been discussing in lecture. They are not in your textbooks. 1. It has been documented for many years that stress can have deleterious effects on health. A recently reported study out of the lab of a Nobel Prize winner has shown a link between stress and the immune system, and also shown that exercise may relate to immune function in a novel way. These studies suggest: a) that mutations in the mTOR gene are linked to lack of exercise; b) that people who exercise produce significantly higher levels of bursicon than those that don’t exercise; c) that stress-induced loss of telomere length is less in people that exercise; d) that exercise-related reductions in breast cancer correlate with increased levels of serum insulin in postmenopausal women; e) all of the above. 2. Sequencing of the human, Neanderthal and chimpanzee genomes have allowed for evolutionary comparisons. Looking at regions that have been both retained and lost in these genomes, several studies over the past five years have found: a) a rapidly-evolving non-coding region on human chromosome 2 that drives human-specific patterns of gene expression in the forelimb; b) a potential enhancer region deleted from the human genome near the androgen receptor gene that correlates to differences in secondary sexual development between primates; c) differences in expression of a tumor-suppressor gene during embryonic brain development that may correlate to brain size; d) all of the above; e) none of the above. 3. During E. coli DNA replication, which of the following is a function of DNA gyrase: a) opens up the replication bubble at the origin of replication; b) unwinds DNA at the replication fork; c) relieves torsional stress by introducing negative supercoils in advance of the replication fork; d) all of the above; e) none of the above. 4. The recognition sites for the restriction enzymes BamHI, XbaI and BglII are 5’-GGATCC-3’, 5’-TCTAGA-3’ and 5’-AGATCT-3’, respectively, where the arrows represent the cut locations for each strand of the palindromic site. Which enzymes leave compatible ends that will facilitate ligation? a) All of these enzymes leave ends that are compatible with ends generated by the others; b) None of the enzymes produce compatible ends; c) Only BamHI and BglII fragments are compatible; d) Only BamHI and XbaI fragments are compatible; e) only BglII and XbaI fragments are compatible. 1 2 3 4 5 6 7 8 9 10 Questions 5-6 pertain to the figure on the right. DNA was prepared from white blood cells isolated from a large human population. These DNAs were individually digested with EcoRI, subjected to electrophoresis and Southern blotting, and the blot was probed with a radioactively labeled cloned DNA sequence. Ten different individuals were analyzed for their patterns. The figure below indicates the results from these ten individuals. 6.4 kb 4.1 kb 2.3 kb 1.6 kb 5. In the diagram on the right, the vertical lines represent EcoRI restriction sites. An asterisk over the site represents a a) polymorphism (presence or absence of the site in 1.6 individuals) in the population. The double arrow above the DNA represents the boundaries of the cloned DNA used c) 1.6 as a probe in the Southern blot analysis. Which of the following is consistent with the arrangement of EcoRI restriction sites in human genomic DNA? e) 1.6 * 2.3 * b) 4.1 * 2.3 2.3 4.1 * 4.1 d) * 1.6 2.3 * * 1.6 4.1 2.3 4.1 6. Which of the above individuals (1-10) would represent individuals that carried chromosomes that were heterozygous for the distribution of restriction sites in this area? a) individuals 1 and 2; b) individuals 3, 4 and 6; c) individuals 2,5, 7 and 10; d) individuals 1, 8 and 9; e) none of the above. The accompanying table shows the presence or absence of seven sequence-tagged sites (STS’s) numbered 1-7, in each of seven BAC clones, designated A-G. A “+” in the table means that PCR primers specific to the STS are able to amplify the sequence from the clone, a “-“ means that the sequence can not be amplified from the clone. STS marker BAC Clone A B C D E F G 1 + - 2 + + + + - 3 + + 4 + + + - 5 + + + + 6 + + + - 7 + 7. The relative order of the STS sites within the cloned genome can be ordered as: a) 4165237; b) 1462537; c) 7352164; d) 7251643; e) 1347256 Questions 8-9 pertain to the following diagram, which represents a region of a DNA sequencing gel produced using the Sanger chain termination method. The individual lanes represent the reaction products from the four separate labeling reactions (A,G,C,T) which were loaded at the top of the gel. 8. As deduced from this experiment, the sequence of the strand being copied (template) in this reaction is: a) 5’-AGACTATCTCTA-3’ b) 5’-ATCTCTATCAGA-3 c) 5 -TAGAGATAGTCT-3’ d) 5- TCTGATAGAGAT-3’ e) 5- GAGTCGCTCTCG-3’ A G C T 9. Which of the following was NOT present in the test tube during the ‘G’ DNA synthesis reaction? a) dATP; b) dGTP; c) ddGTP; d) dTTP; e) all of the above were present in the reaction. Question 10 pertains to the following. Six independently derived mutants are recovered in Neurospora which are all unable to grow unless supplemented with compound Z. The mutants are then grown on minimal media supplemented with one of 6 chemicals all believed to be precursors to compound Z. A summary of the ability of the mutants to grow on media containing these chemicals is indicated below, where a “+” sign indicates growth and a “-” sign indicates no growth. precursors added to minimal medium mutant number A B C D E F Z 1 + + + - - - + 2 + + + + - - + 3 + + + + + - + 4 - - + - - - + 5 - - - - - - + 6 - + + - - - + 10. From the above chart, the enzyme that catalyzes the production of compound A appears to be mutant in strain: a) 1; b) 2; c) 3; d) 4; e) none of the above.