Supplementary Table 1: List of ChrI fragment variants identified in
advertisement
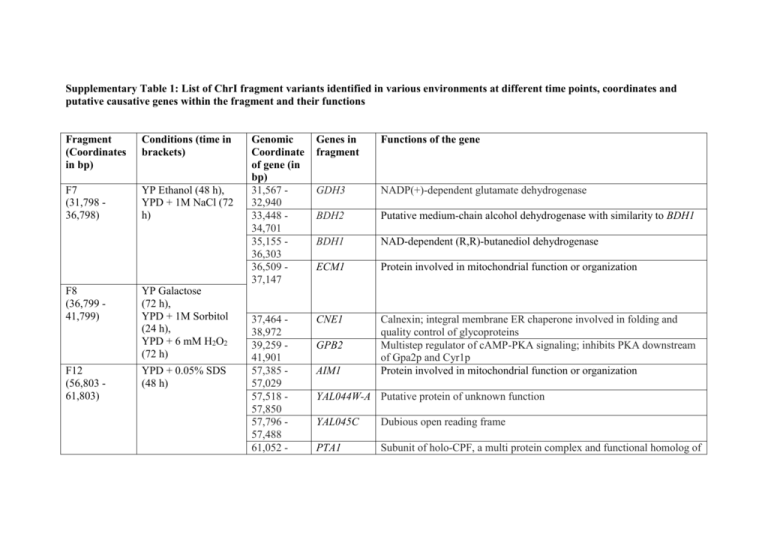
Supplementary Table 1: List of ChrI fragment variants identified in various environments at different time points, coordinates and putative causative genes within the fragment and their functions Fragment (Coordinates in bp) Conditions (time in brackets) F7 (31,798 36,798) YP Ethanol (48 h), YPD + 1M NaCl (72 h) F8 (36,799 41,799) F12 (56,803 61,803) YP Galactose (72 h), YPD + 1M Sorbitol (24 h), YPD + 6 mM H2O2 (72 h) YPD + 0.05% SDS (48 h) Genomic Coordinate of gene (in bp) 31,567 32,940 33,448 34,701 35,155 36,303 36,509 37,147 Genes in fragment Functions of the gene GDH3 NADP(+)-dependent glutamate dehydrogenase BDH2 Putative medium-chain alcohol dehydrogenase with similarity to BDH1 BDH1 NAD-dependent (R,R)-butanediol dehydrogenase ECM1 Protein involved in mitochondrial function or organization 37,464 38,972 39,259 41,901 57,385 57,029 57,518 57,850 57,796 57,488 61,052 - CNE1 Calnexin; integral membrane ER chaperone involved in folding and quality control of glycoproteins Multistep regulator of cAMP-PKA signaling; inhibits PKA downstream of Gpa2p and Cyr1p Protein involved in mitochondrial function or organization GPB2 AIM1 YAL044W-A Putative protein of unknown function YAL045C Dubious open reading frame PTA1 Subunit of holo-CPF, a multi protein complex and functional homolog of 58,695 F13 (61,804 66,804) mammalian CPSF, required for the cleavage and polyadenylation of mRNA and snoRNA 3' ends Protein localized to COPII-coated vesicles; involved in the membrane fusion stage of transport 61,316 62,563 ERV46 62,840 65,404 CDC24 67,520 65,778 87,286 87,752 92,270 87,855 99,697 95,630 99,305 99,868 101,145 00,225 105,872 101,565 CLN3 Guanine nucleotide exchange factor for Cdc42p, required for polarity establishment and maintenance, and mutants have morphological defects in bud formation and shmooing G1 cyclin involved in cell cycle progression SNC1 Vesicle membrane receptor protein (v-SNARE) MYO4 Type V myosin motor involved in actin-based transport of cargos DRS2 Trans-golgi network aminophospholipid translocase (flippase) HRA1 Non-protein-coding RNA MAK16 LTE1 Constituent of 66S pre-ribosomal particles; required for maturation of 25S and 5.8S rRNAs Protein similar to GDP/GTP exchange factors; equired for asymmetric localization of Bfa1p at daughter-directed spindle pole bodies and for mitotic exit at low temperatures 108,551 106,272 PMT2 protein O-mannosyltransferase of the ER membrane YP Galactose (72 h) F18 (86,809 91,809) YP Ethanol (24 h, 48 h) F20 (96,811 10,1811) YP Galactose (48 h, 72 h), YP Fructose (48 h,72 h) F21 (101,812 106,812) YP Maltose (24 h, 48 h) F22 (106,813 - YP Maltose (24 h, 48 h, 72 h) 111,812) F23 (111,814 116,814) F24 (116,815 121,815) F25 (121,816 126,817) F26 (126,817 131,817) YP Galactose (24 h, 48 h, 72 h), YPD + 1M NaCl (72 h) YP Maltose (24 h, 48 h, 72 h), YP Galactose (24 h, 48 h, 72 h), YP Ethanol (24 h,48 h,72 h) YP Maltose (24 h, 48 h), YPD + 4NQO (48 h) YP Maltose (72 h), YP Galactose (24 h, 48 h) YP Galactose (72 h) 110,430 108,877 113,359 110,846 114,615 113,614 114,919 118,314 FUN26 Vacuolar membrane transporter with broad nucleoside selectivity CCR4 Component of the major cytoplasmic deadenylase, CCR4-NOT transcriptional complex Protein required for modification of wobble nucleosides in tRNA ATS1 FUN30 Snf2p family member with ATP-dependent chromatin remodeling activity; has a role in silencing at the mating type locus, telomeres and centromeres 119,541 118,564 120,225 124,295 LDS1 Protein involved in spore wall assembly PSK1 PAS domain-containing serine/threonine protein kinase , co-ordinately regulates protein synthesis and carbohydrate metabolism and storage in response to a unknown metabolite that reflects nutritional status 124,492 124,307 125,069 124,755 124,879 126,786 128,102 126,903 129,019 128,252 129,270 - YAL016C-B Dubious open reading frame YAL016C-A Dubious open reading frame TPD3 NTG1 Regulatory subunit A of the heterotrimeric PP2A complex; equired for cell morphogenesis and transcription by RNA polymerase III DNA N-glycosylase and apurinic/apyrimidinic (AP) lyase SYN8 Endosomal SNARE related to mammalian syntaxin 8 DEP1 Component of the Rpd3L histone deacetylase complex; required for F31 (152,257 157,257) F32 (157,258 162,258) F34 (167,260 172,260) YP Ethanol (24 h, 48 h) 130,487 130,799 131,983 152,257 153,876 154,724 154,065 155,005 156,285 CYS3 diauxic shift-induced histone H2B deposition onto rDNA genes Cystathionine gamma-lyase NUP60 FG-nucleoporin component of central core of the nuclear pore complex ERP1 Member of the p24 family involved in ER to Golgi transport SWD1 Subunit of the COMPASS (Set1C) complex; COMPASS methylates histone H3 on lysine 4 and is required in transcriptional silencing near telomeres Subunit of heterotrimeric Replication Protein A (RPA) which is a highly conserved single-stranded DNA binding protein involved in DNA replication, repair, and recombination 158,619 156,754 RFA1 158,966 159,793 164,187 160,597 166,162 160,238 168,871 166,742 169,375 170,295 171,703 170,396 175,135 172,211 SEN34 Subunit of the tRNA splicing endonuclease; YAR009C Retrotransposon TYA Gag and TYB Pol genes YARCTy1-1 Ty1 element BUD14 Protein involved in bud-site selection ADE1 Required for de novo purine nucleotide biosynthesis KIN3 Nonessential serine/threonine protein kinase CDC12 Protein kinase of the Mitotic Exit Network YP Galactose (72 h) YP Dextrose (72 h)