Chapter 16. Discriminant Analysis and Classification.
advertisement

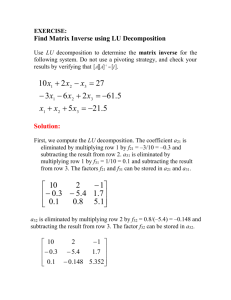
Revised: 2/12/2016 116100998 Chapter 16. Discriminant Analysis. 16:1 What is Discriminant Analysis? Discriminant analysis (both for discrimination and classification) is a statistical technique to organize and optimize: the description of differences among objects that belong to different groups or classes, and the assignment of objects of unknown class to existing classes. For example, we may want to determine what characteristics of the inflorescence best discriminate between two very similar species of grasses, and we may want to create a rule that can be used by others to classify individual plants in the future. Thus, there are two related activities or concepts in discrimination and classification: 1. Descriptive discrimination focuses on finding a few dimensions that combine the originally measured variables and that separate the classes or collections as much as possible. 2. Optimal assignment of new objects, whose real group membership is not known, into one of the existing groups or classes. Discriminant analysis is a method for classifying observations (objects or subjects) into one of two or more mutually exclusive groups; for determining the degree of dissimilarity of observations and groups; and for determining the specific contribution of each independent variable to this dissimilarity. 16:1.1 Elements of DA: One categorical dependent variable (groups or classes); for example, Bromus hordeaceus vs. Bromus madritensis. When we have groups that represent factorial combinations of variables, these have to be “flattened” and considered as a set of groups. For example, if we are trying to identify the species and origin of seeds from 2 species (brma and brho)that may have come from two environments (valley or mountain) we have to create a nominal variable that takes 4 values, one for each possible combination of origin and environment. A set of continuous independent variables that are measured on each individual; for example, length, width, area and perimeter of the seed outline. A set with as many probability density functions (pdf) as there are groups. Each pdf describes the probability of obtaining an object, subject or element from a group that has a particular set of values for the independent variables. For example, the pdf for B. hordeaceus (brho) would tell you the probability of finding a brho seed of any given combination of length, width, area and perimeter. The pdf for B. madritensis (brma) would tell you the probability of finding a brma seed with those same characteristics. Typically, it is assumed that all the pdf’s are the multivariate normal distributions. The equation for the multivariate normal distribution is: f (x) 1 2 p 2 12 e x 1 x 2 1 116100998 Revised: 2/12/2016 where x is the vector of random variables, p is the number of variables or rows in x , is the variance covariance matrix of x , and is the centroid of the distribution. If we were considering only two characteristics, say width and length, the two pdf’s for the two grasses might look like this (after standardizing width and length, simulated data): B. hordeaceus B. madritensis Note that for any combination of length and width there is a positive probability that it be brma, as well as brho. In some areas, the probabilities are clearly different, but in others they are similar. Cutting away the front and left sides of the picture allows us to see better how the two pdf’s interact. 2 116100998 16:1.2 How does DA compare with other methods? 16:1.2.1 with PCA: 16:1.2.2 16:1.2.3 Revised: 2/12/2016 1. DA has X and Y variables, whereas in PCA there is only one set of variables. 2. DA has predetermined groups. 3. Both use the concept of creating new variables that are linear combinations of the original ones. with Cluster Analysis: 1. DA has predetermined groups, and it is used to optimally assign objects of unknown membership to the groups. 2. Cluster analysis is used to generate classifications or taxonomies. 3. In DA, groups are mutually exclusive and exhaustive. All possible groups must be considered, and each object or subject belongs to a single group. This is not the case for all versions of cluster analysis. With MANOVA 1. DA and MANOVA are very similar, and are based on several common theoretical aspects. In fact, DA is accessible through the MANOVA Fit Model personality. 2. Both have categorical X's and continuous Y's (particularly in the discrimination phase of DA). 3. Both use exactly the same canonical variates, separation of SS&CP into between and within groups, etc. 4. The boundary between MANOVA and descriptive DA is not clear-cut in terms of the statistical calculations. The calculations are almost the same. 5. The difference between MANOVA and classification is a clear one in terms of objectives and calculations. Whereas in MANOVA the main question is whether there are significant 3 116100998 Revised: 2/12/2016 differences among groups, in DA the main goal is to develop and use discriminant functions to optimally classify objects into the groups. 16:2 Why and When to use Discriminant Analysis? DA is useful in the following types of situations: Incomplete knowledge of future situations. For example, a population can be classified as being at risk of extinction on the basis of characteristics that were typical of populations that went extinct in the past. A student applying to go to college may have to be classified as likely to succeed or likely to fail based on the characteristics of students who did succeed or fail in the past. The group can be identified, but identification requires destroying the subject or plot. For example, the strength of a rope or a camalot can be measured by stressing it until it breaks. Of course after it breaks we know its strength but cannot use the information on that particular piece, because it no longer exists. The exact species of a seed can be determined by DNA analysis, but after the analysis is done, there is no more seed left to do anything with the information! Unavailable or expensive information. For example, the remains of a human are found and the sex has to be determined. The type of land cover has to be determined for each square km of a large region. Although it would be possible to go to each spot and look at the land cover directly, it would be too expensive. Satellite images can be used and land cover inferred from the spectral characteristics of the reflected radiation. When the goal is classification of objects whose classes are unknown, the analysis proceeds as follows: 1. Obtain a random sample of objects from each class (these are objects whose membership is known). This is known as the "training" or "learning" sample. 2. Measure a series of continuous characteristics on all objects of the training sample and identify any characteristics that are redundant or that really do not help in the discrimination among groups (this can be done by using MANOVA with stepdown analysis, see textbook by Tabachnik and Fidell). This step is not crucial, but can save time, money and increase power of discrimination. 3. Submit the training sample to a DA and obtain a set of discriminant functions. These functions are used implicitly by SAS and JMP, so you do not need to see or know them. The information on these functions is stored in a SAS dataset that is created with an OUTSTAT=file1 option in the PROC DISCRIM statement. In JMP, the discrimination functions can be saved to table columns. 4. In JMP, add a row containing the values of all predictors for an object to the data table. In SAS, create a new SAS dataset (file2) with the characteristics of objects of unknown membership to be classified and submit to another PROC DISCRIM where DATA=file1 and TESTDATA=file2. The same procedure allows a true validation of the classification functions by using a file2 that contains objects of known membership to be classified using only the information on the Y variables and the classification functions developed with an independent dataset. Because the pdf’s of different groups overlap, some classification errors will usually be made, even if the true parameters that describe the pdf's for each group are known. 4 116100998 Revised: 2/12/2016 Figure 16-1. A linear classification rule to determine if people own riding mowers based on their income and lot size. Regardless of the position of the boundary line used for classifying individuals, some individuals will be classified incorrectly. 16:3 Concepts involved in discrimination and classification. A good classification system should have the following characteristics: 1. Use all information available. 2. Make few classification errors. 3. Minimize the negative consequences of making classification errors. Aside from the statistical details, a classification problem has the following elements: 16:3.1 1. Groups or populations. 2. PDF's for each group or population in the X space. 3. Classification rules. 4. Relative sizes of each group. 5. Costs of misclassification. Basic idea Assign the unit with unknown membership to the group that has the maximum likelihood of being the source of the observed vector Xu. Example: 2 urns in random positions. One contains 9 white and 1 black marbles (A) the other contains 1 white and 9 black (B). Blindfolded, you extract one marble from one urn. Where did it come from? The wisest decision rule would be: black B white A However, even knowing all population parameters we will make mistakes. 5 Revised: 2/12/2016 116100998 Outcome A and whiteA A and blackA B and whiteB B and blackB Prob 9/20 1/20 1/20 9/20 Classific. A B A B Error? No Yes Yes No error rate 1/10 The basic classification idea minimizes error rate or cost of errors. The only difference between this example and discriminant analysis is the complexity. The essential theoretical basis is the same. Rule: Assign an individual u to group g if: P(gXu) > P(g’Xu) for all g’ g (for all groups other than g) If we are considering a single continuous variable for the classification, and we have two groups, the decision rule can be depicted with the following Figure. Note that nothing is assumed or said about the specific distribution of the observed variable in each group. Figure 16-2. Classification rule and error rates for two groups when there is a single dimension or variable used for the classification. X is the characteristic measured to classify objects. Population on the left is 1 and the one on the right is 2. P(j|k) is the probability of classifying an object as j given that it is k. 16:3.2 Prior Probabilities Suppose that in the previous example we take 1 urn of type A and 2000 urns of type B. Marbles can come only from 2 groups as before: A or B. Further, suppose that you randomly select a marble from a random urn and it is white. Do you say it came from an urn type A or B? In the previous situation it was clear (almost) that it came from A. As the number of B urns increases, the probability that the white marble came from B also increases. Consider the probability of the event “white marble from B”, call it P(whiteB). P(whiteB) = P(white) P(Bwhite) = P(B) P(whiteB) In general, assume that instead of color you measure a vector Xu on the extracted marble and use g to designate groups. 6 116100998 Revised: 2/12/2016 P(Xug) = P(Xu) P(gXu) = P(g) P(Xug) We are interested in calculating P(gXu) for all g’s, so we can assign Xu (the marble) to the group g with the max P(gXu). P(g) are called prior probabilities, or priors and reflect the probability of getting a unit at random from any g, before we know anything about the unit. (P(g) = pg) 16:3.3 Costs of making errors The cost of incorrectly classifying an individual from group 1 into group 2 may be quite different from the cost of incorrectly putting an individual from group 2 into group 1. A typical example is that of a trial for a serious crime. The truth is not known (perhaps not even to the person on trial). What is the consequence of releasing a guilty subject? What is the consequence of convicting an innocent person? The relative consequences should affect the way in which one weighs the evidence. This is taken into account in discriminant analysis by the decision rule. Note that the following decision rule and figure depict a situation in which we are measuring 2 characteristics of each object, so the whole plane is divided into two regions: R1 : f1 (X) C(1|2) p2 f2 (X) C(2|1) p1 R2 : f1 (X) C( 1|2) p2 f2 (X) C( 2|1) p1 These rules indicate that we should classify the object a in population 1 if the ratio of probabilities (“heights” of the pdf’s) f1(X)/f2(X) is greater than the ratio of the costs of misclassification times the ratio of priors. C (j|k) is the cost of classifying an object from population k into j; pk is the prior probability for population k. 7 116100998 Revised: 2/12/2016 Figure 16-3. Example of decision rule for classification of two populations based on two characteristics. The line partitions the plane of all possible pairs of values (x1, x2) (the "universe" of events ) into two mutually exclusive and comprehensive sets, R1 and R2. This figure shows an unusual shape of the boundary between the two groups, but it is a possible one. 16:4 Model and Assumptions. 16:4.1 Model The model is essentially the same as for MANOVA, except that in DA the categorical variable is always a oneway analysis. Factorial combinations must be “flattened” and viewed as a single set of different groups or treatments. 16:4.2 Assumptions and other issues. 16:4.2.1 Equality of sample size across cells. Inequality of cell sizes is usually not a problem because DA is one-way. Sample size in the smallest group should exceed number of characteristics or variable used for classification (X’s). The procedure is robust against deviations from assumptions if the smallest group has more than 20 cases or observations and there are more than 20 observations per predictor or characteristic used for classification. 16:4.2.2 Multivariate normality. If normality is not achieved, analysis can still be performed for descriptive purposes, but the optimal classification rule cannot be derived through the traditional methods. Alternatively, SAS offers a series of nonparametric alternatives in PROC DISCRIM, or one can use logistic regression. If normality and parametric analysis are desired, transformations of the variables should be tried. 8 116100998 Revised: 2/12/2016 The procedure is robust against lack of normality if sample size gives >20 df in the error of the ANOVA. 16:4.2.3 Independence. As in most analyses, random sampling and independence among observations is essential. In order to assess the adequacy of the sampling, the target populations must be clearly defined. 16:4.2.4 No outliers. Discriminant analysis is sensitive to outliers like MANOVA. Test for outliers using the squared Mahalanobis distance and eliminate those observations with P<0.001. Document and report outlier detection and elimination. Outliers can be detected by using the Mahalanobis distance or its jackknifed version. The procedure is exactly the same as in MANOVA. 16:4.2.5 Homogeneity of Variance-covariance matrices. SAS: test with POOL=TEST option in PROC DISCRIM statement. JMP: obtain the E matrices for each group by using the Fit Model Manova personality. Include all predictors or classification variables in the Y box, and include the grouping variable in the By box. Leave the effects blank. This will give an E matrix and the corresponding partial covariance matrix. The partial covariance matrices can be copied onto a spreadsheet and Box’s M can be calculated using the equation given in the MANOVA notes (note that the vertical bars in the formula for Box’s M represent the determinant of the enclosed matrix, not the absolute value). SAS automatically uses a quadratic discriminant function to account for heterogeneous variance-covariance matrices. JMP uses only linear discriminant functions, so it is not easy to deal with heterogeneity of variance. The linear function can still be used, but the actual error rates will tend to be higher than reported by the model. It is possible to create JMP scripts to calculate quadratic discriminant equations. 16:4.2.6 Linearity Linearity is assumed among all pairs of continuous variables. Discriminant analysis can only incorporate linear relationships among variables. Linearity can be tested by examination of the scatterplots of pairs of variables. If significant non-linearity cannot be fixed by transformations, logistic regression can be used instead of Discriminant analysis. Lack of linearity reduces the power of the tests but does not affect Type I error very much. 16:4.2.7 Multicollinearity or redundant Y's High collinearity among the continuous variables can make matrix inversion very unstable. The degree of collinearity can be checked by examining the tolerance value for each characteristics measured and potentially used for the classification. Tolerance is the inverse of the VIF, or simply 1-R2, where R2 is the coefficient of determination of regressing each one of the continuous variables on the rest. Delete variables whose tolerances are lower than 0.10. 16:4.2.8 Significant differences between groups In order to use the information in the training sample to classify individuals in the future, it is necessary to measure things that discriminate among groups. Variables whose values are significantly different among groups can be detected by performing a MANOVA. Those variables that do not contribute to the differences should be considered for deletion. 16:4.2.9 Generality of classification functions When the true pdf's and prior probabilities are known, as in the urn example above, the exact error rates can be calculated. However, in most real situations, neither the pdf's not the priors are known. Error rates must be estimated from the training sample and, if possible, further validated with independent data. SAS performs two analyses of errors, a re-substitution and a cross-validation. The re-substitution analysis simply applies the classification rule to the training sample. This, of course, underestimates error rates because it is based on the same data used to develop the classification or discrimination function. The cross-validation is also known as the hold-out methods and is a jackknifing procedure. Each observation in the training sample is classified based on a rule obtained without the observation in the data (each observation is "held out," one at a time). The option of performing a true validation is tricky, because if an independent data set with objects of known membership were available, it would not make sense to ignore it for the development of the classification rule. 9 Revised: 2/12/2016 116100998 Do not extrapolate beyond the population sampled. For example, if you desire to determine if your dog is about to attack based on the multivariate characteristics of the barking sounds, you only need to measure your dog, and you should not use the classification function derived for other dogs. 16:5 Obtaining and interpreting output in JMP. Consider an example in which you need to create an automated system to classify seeds of Bromus hordaceus (brho), Bromus madritensis (brma) and Lolium multiflorum. The system is based on imaging techniques that can automatically measure the length, width, area and perimeter of the seed’s “shadow.” You have a sample of seeds for which you know the species with certainty (training or learning sample). The summary statistics for the sample are given in the following table, where linear dimensions are in mm and areas are in mm 2. The measurements were obtained by scanning a piece of paper where the seeds had been glued flat, and by using the “particle analysis” feature of NIH Image 1.62 (image analysis software). Spp. brma lomu brho n area 124 13.5 95 9.4 112 11.0 perim 25.0 14.0 16.3 length 10.7 5.7 6.8 width 1.6 2.1 2.0 sd area sd per 3.4 6.1 2.7 2.7 2.6 2.4 sd len sd wid 1.6 0.4 0.9 0.4 0.8 0.3 In addition to these variables, the ratio of area to perimeter and of width to length were calculated as indices of shape. The data are in the file xmpl_seedDA.jmp. For the purpose of the example, the assumptions are not checked. However, the data show that there is some heterogeneity of variance because brma has more variance, particularly in perimeter and length. The data also includes a few outliers in the brma group, and the variables are highly collinear. These departures are not major, but will tend to increase the error rates relative to what the analysis indicates. The first step is to explore the degree of separation among species in the measured variables. For this, a MANOVA is performed, although this is not a mandatory step. Only the main results are shown here. In any case, DA in JMP is accessed through the Fit Model platform by selecting the Manova personality. The biplot and test details show that the species differ significantly in size and shape of seeds. The characteristics of the seeds show a high degree of collinearity among treatments, as indicated by the facts that they differ almost exclusively in the Canonical 1 direction, and that the first eigenvalue accounts for 97.5% of the explained variance. These elements support the use of seed dimensions and shape to discriminate and classify 10 116100998 Revised: 2/12/2016 seeds of unknown species. In agreement with the results of direct observation of the seeds, it is easier to discriminate between brma and the other two than between lomu and brho. In the same window where the Manova results are displayed by JMP, click on the red triangle to the left of Manova Fit and select “Save Discriminant.” This command will result in the addition of a series of columns to the data table that contain the Mahalanobis distance from each observation to each centroid. 11 116100998 Revised: 2/12/2016 The Column labeled Dist[0] contains the part of the distance that is the same, regardless of group. Each Dist[i] column uses Dist[0] to calculate the distance from the observation to the centroid for group i. The columns labeled Prob[i] contain the posterior conditional probabilities for group membership. They are posterior because they are calculated on the basis of the dimensions of the seeds. They are conditional because the give the probability of membership given that the seed has the observed dimensions. For example, Prob[brho]= 0.29150079 for observation 1 means that there is a probability of 0.29150079 that a seed with the characteristics of that in observation 1 is a brho seed. For the same observation, Prob[lomu]= 0.70846982. This means that on average. out of 100,000 seeds with dimensions equal to those in observation 1, 29,150 will be brho, 70,847 will be lomu, and the rest will be brma. Yet, we would classify all of them as lomu! JMP assumes that all classes are equally likely in the universe of classes, meaning that all prior probabilities are the same and equal to 1/number of classes. If one has information indicating that the classes are not equally frequent, that information can be incorporated in the classification scheme, but that requires a minimum of understanding of the equations used for the classification. These equations and an excellent treatment of the subject can be found in Chapter 11 of Johnson and Wichern (1998). 16:6 Obtaining and interpreting output in SAS. data crops; title 'DA of crop remote sensing data'; input crop $ x1-x4 xvalues $ 10-26; cards; corn 16 27 31 33 corn 15 23 30 30 corn 16 27 27 26 The variable xvalues is simply a label corn 18 20 25 23 that contains the values of all x corn 15 15 31 32 variables, for the purpose of identifying corn 15 32 32 15 the observations. The numbers 10-26 corn 12 15 16 73 tell the program to read the label from soyb 20 23 23 25 positions 10 through 26 in each line. soyb 24 24 25 32 soyb 21 25 23 24 soyb 27 45 24 12 soyb 12 13 15 42 soyb 22 32 31 43 cotton 31 32 33 34 cotton 29 24 26 28 cotton 34 32 28 45 cotton 26 25 23 24 cotton 53 48 75 26 cotton 34 35 25 78 sugarb 22 23 25 42 sugarb 25 25 24 26 sugarb 34 25 16 52 sugarb 54 23 21 54 sugarb 25 43 32 15 sugarb 26 54 2 54 clover 12 45 32 54 clover 24 58 25 34 clover 87 54 61 21 clover 51 31 31 16 clover 96 48 54 62 clover 31 31 11 11 12 Revised: 2/12/2016 116100998 clover clover clover clover clover ; 56 32 36 53 32 13 13 26 8 32 13 27 54 6 62 71 32 32 54 16 proc discrim data=crops outstat=cropstat method=normal pool=test list crossvalidate; class crop; priors prop; id xvalues; var x1-x4; title2 'Using the discriminant function on a test dataset'; run; data test; input crop $ x1-x4 xvalues $ 10-26; corn 16 27 31 33 soyb 21 25 23 24 cotton 29 24 26 28 sugarb 54 23 21 54 clover 32 32 62 16 ; The OUTSTATS option creates a SAS dataset that contains all information for classification of new individuals or samples. METHOD requests either a parametric (multivariate normality assumed) or nonparametric classification rules. Data set used to load the classification information based on the previous analysis. proc discrim data=cropstat testdata=test testou=tout testlist; class crop; testid xvalues; title2 'Classification of test data'; run; New data with observations to be classified. proc print data=tout; title2' output of classification of test data'; run; 13 Revised: 2/12/2016 116100998 Discriminant Analysis 36 Observations 35 DF Total 4 Variables 31 DF Within Classes 5 Classes 4 DF Between Classes Priors are set to be equal to the proportions in the training sample. Class Level Information CROP clover corn cotton soyb sugarb Frequency 11 7 6 6 6 Weight 11.0000 7.0000 6.0000 6.0000 6.0000 Proportion 0.305556 0.194444 0.166667 0.166667 0.166667 Prior Probability 0.305556 0.194444 0.166667 0.166667 0.166667 Discriminant Analysis Within Covariance Matrix Information Covariance Natural Log of the Determinant CROP Matrix Rank of the Covariance Matrix This is an index of the "amount" of clover 4 23.64618 variance in each group and in the corn 4 11.13472 pooled sample. Very large cotton 4 13.23569 negative numbers indicate soyb 4 12.45263 collinearity. sugarb 4 17.76293 Pooled 4 21.30189 Discriminant Analysis Notation: K = P = N = N(i) = V RHO DF Under null hypothesis: Test of Homogeneity of Within Covariance Matrices Number of Groups Number of Variables Total Number of Observations - Number of Groups Number of Observations in the i'th Group - 1 __ N(i)/2 || |Within SS Matrix(i)| = ----------------------------------N/2 |Pooled SS Matrix| _ | 1 = 1.0 - | SUM ----|_ N(i) - Test for homogeneity of variance-covariance matrices among groups. _ 2 1 | 2P + 3P - 1 --- | ------------N _| 6(P+1)(K-1) = .5(K-1)P(P+1) _ _ | PN/2 | | N V | -2 RHO ln | ------------------ | | __ PN(i)/2 | |_ || N(i) _| Homogeneity of variancecovariance is rejected, so a quadratic discriminant formula is used. is distributed approximately as chi-square(DF) Test Chi-Square Value = 98.022966 with 40 DF Prob > Chi-Sq = 0.0001 Since the chi-square value is significant at the 0.1 level, the within covariance matrices will be used in the discriminant function. Reference: Morrison, D.F. (1976) Multivariate Statistical Methods p252. Discriminant Analysis Pairwise Generalized Squared Distances Between Groups 2 _ _ -1 _ _ D (i|j) = (X - X )' COV (X - X ) + ln |COV | - 2 ln PRIOR i j j i j j j 14 Revised: 2/12/2016 116100998 From Generalized Squared Distance to CROP clover corn cotton 26.01743 1320 104.18297 27.73809 14.40994 150.50763 26.38544 588.86232 16.81921 27.07134 46.42131 41.01631 26.80188 332.11563 43.98280 CROP clover corn cotton soyb sugarb Posterior Probability of Membership in each CROP: 2 2 Pr(j|X) = exp(-.5 D (X)) / SUM exp(-.5 D (X)) j k k XVALUES 27 23 27 20 15 32 15 23 24 25 45 13 32 32 24 32 25 48 35 23 25 25 23 43 54 45 58 54 31 48 31 13 13 26 8 32 31 30 27 25 31 32 16 23 25 23 24 15 31 33 26 28 23 75 25 25 24 16 21 32 2 32 25 61 31 54 11 13 27 54 6 62 sugarb 31.40816 25.55421 37.15560 23.15920 21.34645 Resubstitution Results using Quadratic Discriminant Function Generalized Squared Distance Function: Results of 2 _ -1 _ D (X) = (X-X )' COV (X-X ) + ln |COV | - 2 ln PRIOR classifying the j j j j j j objects in the This SAS procedure shows the equations used. 16 15 16 18 15 15 12 20 24 21 27 12 22 31 29 34 26 53 34 22 25 34 54 25 26 12 24 87 51 96 31 56 32 36 53 32 soyb 194.10546 38.36252 52.03266 16.03615 107.95676 33 30 26 23 32 15 73 25 32 24 12 42 43 34 28 45 24 26 78 42 26 52 54 15 54 54 34 21 16 62 11 71 32 32 54 16 From CROP corn corn corn corn corn corn corn soyb soyb soyb soyb soyb soyb cotton cotton cotton cotton cotton cotton sugarb sugarb sugarb sugarb sugarb sugarb clover clover clover clover clover clover clover clover clover clover clover training sample. Classified Posterior Probability of Membership in CROP: into CROP clover corn cotton soyb sugarb corn 0.0152 0.9769 0.0000 0.0000 0.0079 corn 0.0015 0.9947 0.0000 0.0000 0.0038 corn 0.0023 0.9825 0.0000 0.0000 0.0152 corn 0.0107 0.9793 0.0000 0.0020 0.0079 corn 0.0061 0.9831 0.0000 0.0000 0.0108 corn 0.0070 0.9472 0.0000 0.0000 0.0458 corn 0.0013 0.9987 0.0000 0.0000 0.0000 soyb 0.0097 0.0039 0.0000 0.9772 0.0092 soyb 0.0258 0.0000 0.0014 0.7557 0.2171 soyb 0.0062 0.0000 0.0002 0.9868 0.0068 soyb 0.0105 0.0000 0.0000 0.9807 0.0088 soyb 0.0131 0.0000 0.0000 0.9862 0.0006 soyb 0.0270 0.0000 0.0000 0.9729 0.0001 cotton 0.0285 0.0000 0.9592 0.0032 0.0092 cotton 0.0357 0.0000 0.7796 0.0004 0.1842 cotton 0.0519 0.0000 0.9363 0.0000 0.0118 cotton 0.0123 0.0000 0.9354 0.0444 0.0080 cotton 0.0093 0.0000 0.9907 0.0000 0.0000 cotton 0.0044 0.0000 0.9956 0.0000 0.0000 soyb * 0.0457 0.0000 0.0000 0.8056 0.1487 cotton * 0.0204 0.0000 0.4968 0.4326 0.0503 sugarb 0.0747 0.0000 0.0000 0.0000 0.9253 sugarb 0.2737 0.0000 0.0000 0.0000 0.7263 sugarb 0.2010 0.0000 0.0000 0.0119 0.7871 sugarb 0.0094 0.0000 0.0000 0.0000 0.9906 clover 1.0000 0.0000 0.0000 0.0000 0.0000 clover 0.9704 0.0000 0.0000 0.0001 0.0296 clover 1.0000 0.0000 0.0000 0.0000 0.0000 clover 0.9884 0.0000 0.0000 0.0000 0.0116 clover 1.0000 0.0000 0.0000 0.0000 0.0000 clover 1.0000 0.0000 0.0000 0.0000 0.0000 sugarb * 0.2605 0.0000 0.0000 0.0000 0.7395 sugarb * 0.2987 0.0000 0.0000 0.0000 0.7013 clover 1.0000 0.0000 0.0000 0.0000 0.0000 clover 1.0000 0.0000 0.0000 0.0000 0.0000 clover 1.0000 0.0000 0.0000 0.0000 0.0000 * Misclassified observation 15 Revised: 2/12/2016 116100998 Discriminant Analysis Classification Summary for Calibration Data: WORK.CROPS Resubstitution Summary using Quadratic Discriminant Function Generalized Squared Distance Function: 2 _ -1 _ Re-substitution D (X) = (X-X )' COV (X-X ) + ln |COV | - 2 ln PRIOR summary j j j j j j Posterior Probability of Membership in each CROP: 2 2 Pr(j|X) = exp(-.5 D (X)) / SUM exp(-.5 D (X)) j k k From CROP clover corn cotton soyb sugarb Total Percent Priors Rate Priors Number of Observations and Percent Classified into CROP: clover corn cotton soyb sugarb 9 0 0 0 2 81.82 0.00 0.00 0.00 18.18 0 7 0 0 0 0.00 100.00 0.00 0.00 0.00 0 0 6 0 0 0.00 0.00 100.00 0.00 0.00 0 0 0 6 0 0.00 0.00 0.00 100.00 0.00 0 0 1 1 4 0.00 0.00 16.67 16.67 66.67 9 7 7 7 6 25.00 19.44 19.44 19.44 16.67 0.3056 0.1944 0.1667 0.1667 0.1667 Error Count Estimates for CROP: clover corn cotton 0.1818 0.0000 0.0000 0.3056 0.1944 0.1667 soyb 0.0000 0.1667 sugarb 0.3333 0.1667 Cross-validation Summary using Quadratic Discriminant Function Generalized Squared Distance Function: 2 _ -1 _ D (X) = (X-X )' COV (X-X ) + ln |COV | - 2 ln PRIOR j (X)j (X)j (X)j (X)j j Posterior Probability of Membership in each CROP: 2 2 Pr(j|X) = exp(-.5 D (X)) / SUM exp(-.5 D (X)) j k k From CROP clover corn cotton soyb sugarb Total Percent Priors Number of Observations and Percent Classified into CROP: clover corn cotton soyb sugarb 9 0 0 0 2 81.82 0.00 0.00 0.00 18.18 3 2 0 0 2 42.86 28.57 0.00 0.00 28.57 3 0 2 0 1 50.00 0.00 33.33 0.00 16.67 3 0 0 2 1 50.00 0.00 0.00 33.33 16.67 3 0 1 1 1 50.00 0.00 16.67 16.67 16.67 21 2 3 3 7 58.33 5.56 8.33 8.33 19.44 0.3056 0.1944 0.1667 0.1667 0.1667 Total 11 100.00 7 100.00 6 100.00 6 100.00 6 100.00 36 100.00 Total 0.1111 Crossvalidation summary. Total 11 100.00 7 100.00 6 100.00 6 100.00 6 100.00 36 100.00 16 Revised: 2/12/2016 116100998 Rate Priors Error Count Estimates for CROP: clover corn cotton 0.1818 0.7143 0.6667 0.3056 0.1944 0.1667 soyb 0.6667 0.1667 sugarb 0.8333 0.1667 Total 0.5556 Validation on Discriminant Analysis Classification Results for Test Data: WORK.TEST new data set. Classification Results using Quadratic Discriminant Function (Test data). Generalized Squared Distance Function: Posterior Probability of Membership in each CROP: 2 _ -1 _ 2 2 D (X) = (X-X )' COV (X-X ) + ln |COV | Pr(j|X) = exp(-.5 D (X)) / SUM exp(-.5 D (X)) j j j j j j k k Posterior Probability of Membership in CROP: XVALUES 16 21 29 54 32 27 25 24 23 32 31 23 26 21 62 33 24 28 54 16 From CROP corn soyb cotton sugarb clover Classified into CROP corn soyb cotton sugarb clover clover 0.0152 0.0062 0.0357 0.2737 1.0000 corn 0.9769 0.0000 0.0000 0.0000 0.0000 cotton 0.0000 0.0002 0.7796 0.0000 0.0000 soyb 0.0000 0.9868 0.0004 0.0000 0.0000 sugarb 0.0079 0.0068 0.1842 0.7263 0.0000 Classification Summary using Quadratic Discriminant Function Generalized Squared Distance Function: Posterior Probability of Membership in each CROP: 2 _ -1 _ D (X) = (X-X )' COV (X-X ) + ln |COV | j j j j j Number of Observations and Percent Classified into CROP: clover corn cotton soyb sugarb 1 0 0 0 0 100.00 0.00 0.00 0.00 0.00 0 1 0 0 0 0.00 100.00 0.00 0.00 0.00 0 0 1 0 0 0.00 0.00 100.00 0.00 0.00 0 0 0 1 0 0.00 0.00 0.00 100.00 0.00 0 0 0 0 1 0.00 0.00 0.00 0.00 100.00 1 1 1 1 1 20.00 20.00 20.00 20.00 20.00 0.3056 0.1944 0.1667 0.1667 0.1667 From CROP clover corn cotton soyb sugarb Total Percent Priors Rate Priors 2 2 Pr(j|X) = exp(-.5 D (X)) / SUM exp(-.5 D (X)) j k k Error Count Estimates for CROP: clover corn cotton 0.0000 0.0000 0.0000 0.3056 0.1944 0.1667 OBS CROP X1 X2 X3 X4 1 2 3 4 5 corn soyb cotton sugarb clover 16 21 29 54 32 27 25 24 23 32 31 23 26 21 62 33 24 28 54 16 XVALUES 16 21 29 54 32 27 25 24 23 32 31 23 26 21 62 33 24 28 54 16 soyb 0.0000 0.1667 sugarb 0.0000 0.1667 Total 1 100.00 1 100.00 1 100.00 1 100.00 1 100.00 5 100.00 Total 0.0000 CLOVER CORN COTTON SOYB SUGARB _INTO_ 0.01518 0.00624 0.03569 0.27373 1.00000 0.97691 0.00003 0.00000 0.00000 0.00000 0.00000 0.00017 0.77963 0.00000 0.00000 0.00000 0.98678 0.00043 0.00000 0.00000 0.00791 0.00678 0.18425 0.72627 0.00000 corn soyb cotton sugarb clover 17