Stitching oligos together for fragment synthesis
advertisement

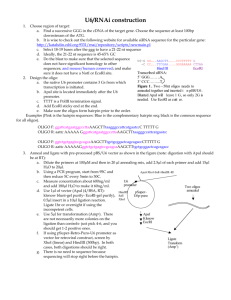
Gene Synthesis from Oligos This method works for stitching together oligos to make longer fragments. The basic idea is that you split your desired DNA into fragments that are made by annealing complementary oligos up to 60 nt each (to avoid larger synthesis scale and higher price per bp). These fragments are made to have overhangs so they can be ligated together. The ability to make long overhangs facilitates ligation and ensures the correct order. Since oligos are not 5’-phosphorylated, you need to kinase them first. 1. Design your sequence. Make a GCK file of the DNA you want to build. If it’s a coding sequence, you should pay attention to codon bias. There is a site at http://gcua.schoedl.de/ that lets you see how a sequence compares to genome-wide codon bias in any of several organisms. Select each codon versus usage table. You’ll get the screen below. Type in a sequence name. Select “originating organism”. Paste in the coding sequence from a gene of interest. In the “Codon usage table to apply” window, select an organism. Click the “frequency” radio button. Click “Submit”. The resulting graph looks something like this. If you used default colors, red is your input sequence and black is all genes. In this case, we might notice that AGA is a bad codon to use for Arg - best to use CGC. Last edited 08/22/2011 by Jeff 2. Break sequence into overlapping oligos. For this step, I find it easiest to print out a nice sequence from GCK, in a largish font, bold, with lots of space between lines. For a sequence of 240 bp, I just cut into 4x60 bp fragments. Adjust as necessary. You need to overlap fragments so they’ll anneal. I’ve done this with InFusion, where I wanted 15-nt 5’ overhangs at the ends, so it seemed easiest to have similar overhangs between fragments, as in the diagram below. The first three (A, B, and C) were 60-nt each, so when annealed I got 15-nt overhang-45 bp duplex-15 nt overhang. 3. Anneal oligos. Resuspend oligos as usual, in TE at 50 pmol / μl. Mix equal amounts of each oligo. You need 6 μl for the kinase reaction, so you could just try 3 and 3, but I usually do 10 and 10. Run any oligo annealing program on a PCR machine. 4. Kinase oligos. For the kinase reaction, mix: 38 μl H2O 6 μl annealed oligos 5 μl T4 PNK buffer 1 μl T4 polynucleotide kinase enzyme Incubate for 1 hr at 37°C. 4. Ligate oligos. Mix all of your phosphorylated oligos together and add enough 10X ligase buffer for it to be 1X, plus 1 μl T4 DNA ligase. Ligate several hours to overnight at room temp. Long overhangs will anneal stably enough at room temp for the ligation to work well. 16°C will only slow down the enzyme activity. 5. Clone! Use your ligated fragment in your desired cloning reaction. You can run some on a gel to check ligation - try 20 μl. You can even cut out the fully-ligated fragment for purification. I’ve done InFusion cloning with 20 μl ligated fragment plus cut vector and had success. Last edited 08/22/2011 by Jeff