U6/RNAi construction
advertisement
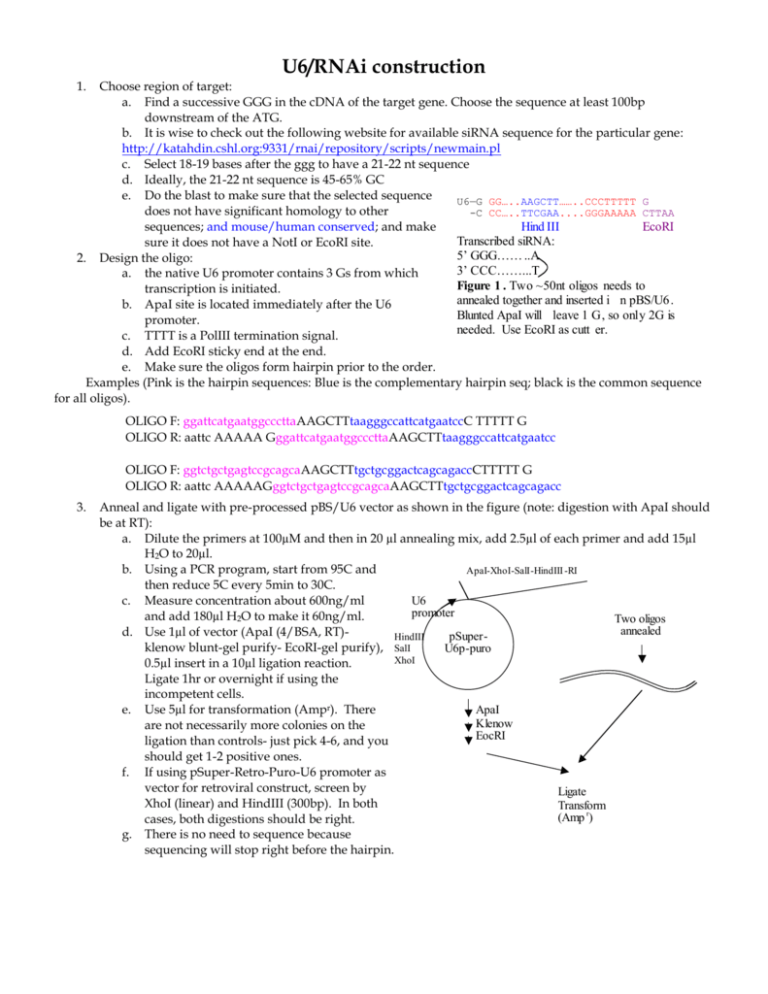
U6/RNAi construction 1. Choose region of target: a. Find a successive GGG in the cDNA of the target gene. Choose the sequence at least 100bp downstream of the ATG. b. It is wise to check out the following website for available siRNA sequence for the particular gene: http://katahdin.cshl.org:9331/rnai/repository/scripts/newmain.pl c. Select 18-19 bases after the ggg to have a 21-22 nt sequence d. Ideally, the 21-22 nt sequence is 45-65% GC e. Do the blast to make sure that the selected sequence U6—G GG…..AAGCTT……..CCCTTTTT G does not have significant homology to other -C CC…..TTCGAA....GGGAAAAA CTTAA Hind III EcoRI sequences; and mouse/human conserved; and make Transcribed siRNA: sure it does not have a NotI or EcoRI site. 5’ GGG…… ..A 2. Design the oligo: 3’ CCC……...T a. the native U6 promoter contains 3 Gs from which Figure 1 . Two ~50nt oligos needs to transcription is initiated. annealed together and inserted i n pBS/U6 . b. ApaI site is located immediately after the U6 Blunted ApaI will leave 1 G, so only 2G is promoter. needed. Use EcoRI as cutt er. c. TTTT is a PolIII termination signal. d. Add EcoRI sticky end at the end. e. Make sure the oligos form hairpin prior to the order. Examples (Pink is the hairpin sequences: Blue is the complementary hairpin seq; black is the common sequence for all oligos). OLIGO F: ggattcatgaatggcccttaAAGCTTtaagggccattcatgaatccC TTTTT G OLIGO R: aattc AAAAA GggattcatgaatggcccttaAAGCTTtaagggccattcatgaatcc OLIGO F: ggtctgctgagtccgcagcaAAGCTTtgctgcggactcagcagaccCTTTTT G OLIGO R: aattc AAAAAGggtctgctgagtccgcagcaAAGCTTtgctgcggactcagcagacc 3. Anneal and ligate with pre-processed pBS/U6 vector as shown in the figure (note: digestion with ApaI should be at RT): a. Dilute the primers at 100µM and then in 20 µl annealing mix, add 2.5µl of each primer and add 15µl H2O to 20µl. b. Using a PCR program, start from 95C and ApaI-XhoI-SalI-HindIII -RI then reduce 5C every 5min to 30C. U6 c. Measure concentration about 600ng/ml promoter and add 180µl H2O to make it 60ng/ml. Two oligos annealed d. Use 1µl of vector (ApaI (4/BSA, RT)HindIII pSuperklenow blunt-gel purify- EcoRI-gel purify), SalI U6p-puro XhoI 0.5µl insert in a 10µl ligation reaction. Ligate 1hr or overnight if using the incompetent cells. ApaI e. Use 5µl for transformation (Ampr). There Klenow are not necessarily more colonies on the EocRI ligation than controls- just pick 4-6, and you should get 1-2 positive ones. f. If using pSuper-Retro-Puro-U6 promoter as vector for retroviral construct, screen by Ligate XhoI (linear) and HindIII (300bp). In both Transform (Amp r) cases, both digestions should be right. g. There is no need to sequence because sequencing will stop right before the hairpin.