Computational structure characterization tools in application to
advertisement

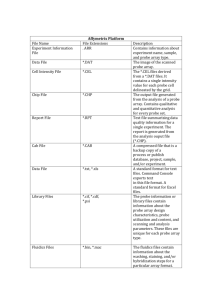
Computational structure characterization tools in application to ordered and disordered porous materials Lev Sarkisov and Alex Harrison Institute for Materials and Processes, School of Engineering, University of Edinburgh, EH9 3JL, UK Abstract In this article we present a set of computational tools for systematic characterization of ordered and disordered porous materials. These tools include calculation of the accessible surface area and geometric pore size distribution, analysis of the structure connectivity and percolation analysis of the porous space. We briefly discuss the algorithms behind these calculations. To demonstrate the capabilities of the tools and the type of insights that can be gained from their application we consider a series of case studies. These case studies include small molecular fragments, several crystalline metal-organic materials, and variants of these materials with induced defects and disorder in their structure. The simulation package is available upon request. Supplemental Information I. Molecular visualizations of IRMOF-1, 10, 16 materials (from left to right, respectively). IRMOF-1 and IRMOF-10 feature two types of cavities, depending on the orientation of the paddlewheel linkers. The tangent orientation (with respect to the inscribed sphere) leads to a cavity with a larger characteristic size of the pore as measured by the inscribed sphere (yellow sphere). The inward orientation of the paddlewheels leads to a smaller effective size of the cage (blue sphere). IRMOF-16 features only one type of cage as a result of the aromatic rings within the linker being in various orientations. II. Examples of the input files for the computational tools: 1 A. Accessible surface area DREIDING.atoms IRMOF.xyz 3.314 500 25.83200 90.00000 0.593 pot 25.83200 90.00000 ! file containing the atom types ! file containing the coordinates of adsorbent ! probe size in Å ! number of trials 25.83200 ! dimensions of the unit cell 90.00000 ! unit cell angles yz, xz, yx ! crystal density in g / cm3 ! type of surface: ! hs - probed with a hard sphere probe ! pot - probed at the potential minimum distance (2**1/6 sigma) 1938098 1 ! random number seed ! surface visualization option (1: yes; 0: no) B. Geometric pore size distribution DREIDING.atoms ! file containing the atom types IRMOF.xyz ! file containing the coordinates of adsorbent 5000 ! number of trials 0.2 ! lower limit on the diameter of the probe 0.2 ! increment of the probe diameter 40 ! maximum probe diameter 25.83200 25.83200 25.83200 ! dimensions of the unit cell 90.00000 90.00000 90.00000 ! unit cell angles yz, xz, yx 328420 ! random seed number 1 ! logical variable: 1 if you want to save coordinates of the probes of specific diameter, 0 - no 13.0, 14.0 ! probe diameter range within which to save the coordinates C. Structure connectivity IRMOF.xyz ! file containing the coordinates of adsorbent 25.83200 25.83200 25.83200 ! dimensions of the unit cell 90.000 90.000 90.000 ! unit cell angles yz, xz, yx 2.00 ! distance between the two connected particles Å D. Pore space connectivity DREIDING.atoms IRMOF.xyz 4.0 ! file containing the atom types ! file containing the coordinates of adsorbent ! lower limit on the probe size 2 0.50 7.00 0.5 25.83200 90.000 25.83200 90.000 ! probe size increment ! upper probe size limit ! grid spacing 25.83200 ! dimensions of the unit cell 90.000 ! unit cell angles yz, xz, yx Dm. Pore space connectivity for a molecular probe DREIDING.atoms IRMOF.xyz Benzene.mol 0.5 25.83200 25.83200 90.000 90.000 21908391 ! file containing the atom types ! file containing the coordinates of adsorbent ! file containing the geometry of the probe ! grid spacing 25.83200 ! dimensions of the unit cell 90.000 ! unit cell angles yz, xz, yx ! random seed III. Computer visualization of the percolated pathway accessible to a benzene molecule in IRMOF-1. 3