Thesis
advertisement

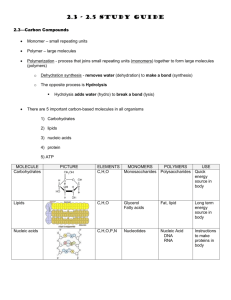
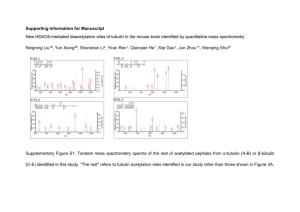
PhD Thesis proposal form Discipline Biology Doctoral School Gènes, Génomes, Cellules Thesis subject title: Regulatory networks of microtubule posttranslational modifications Laboratory name and web site: Institut Curie, Department of Signalling, Neurobiology and Cancer Research team Janke: “The role of tubulin polymodifications in microtubule functions" Web: http://perso.curie.fr/Carsten.Janke/ PhD supervisor (contact person): Name: Carsten JANKE Position: Senior group leader Institut Curie (Research Director CNRS) email: Carsten.Janke@curie.fr Phone number: +33 1 69863127 Thesis proposal: Microtubules are key components of the eukaryotic cytoskeleton involved in a multitude of essential cellular functions. In the past decades, thorough studies have elucidated the biophysical properties of microtubules, their interactions with multiple microtubule-associated proteins and molecular motors, as well as many specialized cellular functions of the microtubule network. While many of the basic functions and properties of microtubules have been studied in great detail, microtubules have generally been considered as homogeneous macromolecular assemblies. This is contrasted by the obvious existence of microtubule identities in cells, which allow microtubules to acquire specific properties for functional specialisation. Our group investigates an emerging regulatory mechanism that could play a key role in generating distinct microtubule identities, the posttranslational modifications of tubulin1. We are interested in the specific role of the modifications polyglutamylation and polyglycylation in the cell cycle control, neuronal differentiation and function as well as in ciliary functions. Previous work has demonstrated that polyglutamylation is enriched on neuronal microtubules2. Very high levels of polyglutamylation are present on axonemal microtubules in cilia and flagella3-5, and also on centrioles, the core components of centrosomes and basal bodies6. Interphase microtubules of most other mammalian cell types carry low levels of polyglutamylation, whereas microtubules of the mitotic spindle and the midbody show elevated polyglutamylation levels7, indicating a possible role of this modification in cell cycle control. Polyglycylation is completely absent from cytosolic microtubules and labels exclusively the axonemes of most cilia and flagella8,9, which could point to a very specific role of this modification in ciliary assembly and functions. The research on these modifications was for many years hampered by the lack of solid functional studies, which could not been carried out since the modifying enzymes had remained unknown. Our group has identified most of the enzymes involved in the generation and removal of polyglutamylation and polyglycylation10-14. Using these enzymes, our group is now using systematic functional studies to understand the roles of polyglutamylation and polyglycylation in microtubule functions. To understand the complexity of the modifications from a molecular to an organism level, we have developed a systems research approach ranging from biochemistry to cell and mouse biology, and we are also investigating links between tubulin modifications and diseases, such as cancer, neurodegeneration and ciliopathies. Despite the fact that we are now able to directly address the role of tubulin polymodifications in a number of microtubule functions, we are still missing insight into the regulatory events that localize, activate or inactive the modifying enzymes. When TTLL proteins are expressed in different cell lines and primary cell culture, the overexpressed proteins rarely show strong localizations within the cells. Some enzymes accumulate at the centrosomes, whereas some localize specifically into cilia15. Only in rare cases, TTLLs localize to cytoplasmic MTs, indicating that those enzymes contain a MT binding motif. The general conclusion from these observations is that most TTLL proteins alone cannot efficiently localize to specific microtubules. It is therefore perceivable that binding partners could exist that guide TTLLs to specific microtubule localizations in cells in order to specifically modify these microtubules. The present PhD project will focus on the mechanisms that regulate the modifications of microtubules in different physiological contexts. The goal is to identify binding partners for several TTLL enzymes to study the mechanisms that specify the functions of different TTLLs within cells. A first insight into how interacting proteins could act on TTLLs has been found for the TTLL1 complex. The biochemical purification of this enzyme revealed four binding partners. One of the subunits, PGs1, can autonomously localize to the pericentriolar space when overexpressed in cells. As none of the other proteins of this complex localized specifically within the cells, this suggested that PGs1 is the localization subunit of the TTLL1 complex10,16. All TTLLs are expressed at very low levels, even in cells with high levels of tubulin modifications, such as neurons or ciliated cells. We have developed specific antibodies for each of the TTLLs, and have never been able to detect an endogenous protein with them, neither in immunohistochemistry nor in immuno blots. In contrast, when the enzymes are biochemically enriched, some antibodies detected the endogenous proteins10. Following these observations, we will isolate TTLL binding partners by purifying endogenous protein complexes from native sources, instead of overexpressing the TTLL enzymes for pull-down experiments. The rationale is that binding partners might be equally low expressed and might not associate efficiently to the overexpressed TTLLs. Biochemical purification methods could be partially deduced from our experience with the purification of the TTLL1 complex16. A final step in the purification could be an affinity purification using our specific antibodies for TTLLs. The purified proteins will be analysed by mass spectrometry, and the function of each novel component will be tested by localization, binding and functional assays in cultured cells. Depending on the identity of the interacting proteins, follow-up functional studies will be designed. For instance, localization subunits of TTLL enzymes could be used to study the dynamic subcellular distribution of the modifying enzymes and their protein complexes during the cell cycle, neuronal differentiation or ciliogenesis. It is equally possible that some of the identified interacting proteins will link the modifying enzymes to signalling pathways. In this case, the role of this pathway in the activity and cellular functions of the proteins will be studied. Finally, we also expect that we could identify new substrates of the modifying enzymes, which could also give rise to functional studies on their role in cells. Taken together, the PhD project in our lab aims to insert the newly discovered enzymes for tubulin polyglutamylation and polyglycylation into the regulatory landscape of cells and organisms. The project is based on established methods in our lab, and challenges new frontiers at the same time. We have a solid network of national and international collaborators that will assist us in our project. The Institut Curie is an internationally recognised centre of excellence, hosting under the same roof basic research ranging from physics over cell biology, developmental biology to cancer research. Moreover, the presence of clinicians at the institute gives researchers direct access to clinical research. References: 1. Janke, C. & Bulinski, J.C. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol 12, 773-786 (2011). 2. Audebert, S. et al. Developmental regulation of polyglutamylated alpha- and beta-tubulin in mouse brain neurons. J Cell Sci 107, 2313-2322 (1994). 3. Lechtreck, K.F. & Geimer, S. Distribution of polyglutamylated tubulin in the flagellar apparatus of green flagellates. Cell Motil Cytoskeleton 47, 219-235 (2000). 4. Million, K. et al. Polyglutamylation and polyglycylation of alpha- and beta-tubulins during in vitro ciliated cell differentiation of human respiratory epithelial cells. J Cell Sci 112, 4357-4366 (1999). 5. Schneider, A., Plessmann, U. & Weber, K. Subpellicular and flagellar microtubules of Trypanosoma brucei are extensively glutamylated. J Cell Sci 110 ( Pt 4), 431-437 (1997). 6. Bobinnec, Y. et al. Glutamylation of centriole and cytoplasmic tubulin in proliferating nonneuronal cells. Cell Motil Cytoskeleton 39, 223-232 (1998). 7. Regnard, C., Desbruyeres, E., Denoulet, P. & Eddé, B. Tubulin polyglutamylase: isozymic variants and regulation during the cell cycle in HeLa cells. J Cell Sci 112, 4281-4289 (1999). 8. Bré, M.H. et al. Axonemal tubulin polyglycylation probed with two monoclonal antibodies: widespread evolutionary distribution, appearance during spermatozoan maturation and possible function in motility. J Cell Sci 109, 727-738 (1996). 9. Plessmann, U. & Weber, K. Mammalian sperm tubulin: an exceptionally large number of variants based on several posttranslational modifications. J Protein Chem 16, 385-390 (1997). 10. Janke, C. et al. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science 308, 1758-1762 (2005). 11. van Dijk, J. et al. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol Cell 26, 437-448 (2007). 12. Rogowski, K. et al. Evolutionary divergence of enzymatic mechanisms for posttranslational polyglycylation. Cell 137, 1076-1087 (2009). 13. Wloga, D. et al. TTLL3 Is a tubulin glycine ligase that regulates the assembly of cilia. Dev Cell 16, 867-876 (2009). 14. Rogowski, K. et al. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell 143, 564-578 (2010). 15. van Dijk, J. et al. Polyglutamylation Is a Post-translational Modification with a Broad Range of Substrates. J Biol Chem 283, 3915-3922 (2008). 16. Regnard, C. et al. Characterisation of PGs1, a subunit of a protein complex co-purifying with tubulin polyglutamylase. J Cell Sci 116, 4181-4190 (2003). Publications of the laboratory in the field: Janke C, Bulinski JC. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol 2011, 12: 773-786. Rogowski K, van Dijk J, Magiera MM, Bosc C, Deloulme J-C, Bosson A, Peris L, Gold ND, Lacroix B, Grau MB, Bec N, Larroque C, Desagher S, Holzer M, Andrieux A, Moutin M-J, Janke C. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell 2010, 143: 564-578. Lacroix B, van Dijk J, Gold ND, Guizetti J, Aldrian-Herrada G, Rogowski K, Gerlich DW, Janke C. Tubulin polyglutamylation stimulates spastin-mediated microtubule severing. J Cell Biol 2010, 189: 945-954. Rogowski K, Juge F, van Dijk J, Wloga D, Strub J-M, Levilliers N, Thomas D, Bre M-H, Van Dorsselaer A, Gaertig J, Janke C. Evolutionary divergence of enzymatic mechanisms for posttranslational polyglycylation. Cell 2009, 137: 1076-1087. van Dijk J, Rogowski K, Miro J, Lacroix B, Eddé B, Janke C. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol Cell 2007, 26: 437-448. Specific requirements to apply, if any: We are looking for creative, independent candidates with a strong background in biology and biochemistry. The candidate should be familiar with basic techniques such as molecular cloning, protein purification, protein analysis, cell culture, light microscopy. Knowledge in mouse biology would be an advantage. The laboratory language is English.