Lab 2 GPC - Iowa State University
advertisement

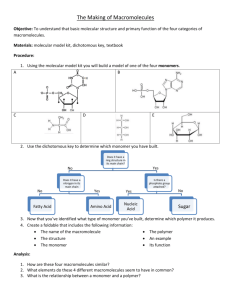
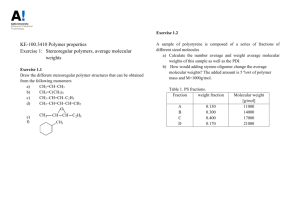
Lab 2: GPC 1. Introduction Gel permeation chromatography (GPC), more correctly termed size exclusion chromatography, is a separation method, which is widely used for estimating the molecular-weight distributions of polymers. The separation takes place in a chromatographic column filled with gel beads that are rigid, highly porous and non-ionic. Highly cross-linked porous polystyrene and porous glass are preferred column-packing materials.1 In a typical GPC experiment, a dilute polymer solution is introduced into the GPC column in which an appropriate solvent is flowing. A GPC column does not separate according to mass, but rather via other mechanisms. As the dissolved polymer molecules flow through the porous beads, they can diffuse into the internal pore structure of the gel to an extent depending on their size and the pore size distribution of the gel. The largest polymer molecules of the solute cannot penetrate the pores within the cross-linked gel beads, and thus elute first (their retention volume is smaller). Smaller polymer molecules of the solute are retained within the gel beads. The larger the molecule, therefore, the less time it spends inside the gel, and the sooner it flows through the column. The different molecular species are eluted (hence called elution time) from the column in order of their molecular size as distinguished from their molecular weight, the largest emerging first (Figure 2.1).2 Once the polymer molecules are separated and come out from column, at least one detector is needed to tell what and how many polymer molecules are at certain time (elution time). The typical detectors are RI (Differential Refractive Index), UV (ultraviolet) and LS (light scattering detector). Some not-so-common detectors can also be found in the GPC systems, such as capillary viscometer detector. 1) The most common RI detector in use today is based on the deflection of a beam of light as it passes through a dual compartment flow cell. One side of the compartment contains the Fred W. Billmeyer, JR. “Textbook of Polymer Science”, Third edition, A willeyInterscience Publication. 2 Jan F. Rabek, “Experimental Methods in Polymer Chemistry”, Physical Principles and Applications, A Wiley-Interscience Publication. 1 1 reference solvent of refractive index n0, which is static during the measurement process. The other side contains the sample solution, i.e., the column eluent, having refractive index n. The differential signal will give the concentration of eluent. In order to use the RI detector quantitatively, the calibration constant must be determined. 2) UV detector is similar to a UV spectrometer, but only detects the UV absorption at the certain wavelength, for our system, 254 nm. The absorption is proportional to the concentration of eluent. 3) Light scattering detector is based on the fact that intensity of light scattered by a polymer molecule is proportional to the square of its mass. LS can be used to give the 'true' molecular weight data for polymer samples, additionally, the information on differences in structure (or composition) between samples. Light scattering detector works with at least one IR detector, which will give the concentration information that LSD needed. GPC is extremely valuable for both analytic and preparative work with a wide variety of systems ranging from low to very high molecular weights. The method can be applied to a wide variety of solvents and polymers depending on the types of gel used. With polystyrene gels, relatively non-polar polymers can be measured in solvents such as tetrahydrofuran, toluene, or (at high temperature) dichlorobenzene. With porous glass gels, more polar systems, including aqueous solvents, can be used. A few milligram of sample is sufficient for analytical work, which could be completed in as short as a few minutes using modern high-pressure, high-speed equipment. Porous Gel Bead Polymer Molecule Figure 2.1. Principle of the separation of molecules according to size by gel permeation chromatography3 3 ERS Department, Trent University, 2 Earlier, it was said that GPC is used to estimate the molecular weight distribution of polymers, including: weight average molecular weight (Mw), number average molecular weight calculation (Mn), polydispersity index (PDI) and average degree of polymerization ( DP ). Theoretically, this information could be obtained using the formulae listed below as Equation 2.1 through Equation 2.4. In these equations, N refers to the number of polymer chains having molecular weight M and for DP calculation, a molecular weight of 104 Daltons for polystyrene will be used. These values are essential in polymer characterization. Molecular weight (calculated as Mn or Mw) of a polymer, for example, is related directly to a polymer’s physical properties. While Mn represents the number average molecular weight, Mw represents the weight average molecular weight. The ratio of the two gives information about the distribution of the chain lengths within a polymer, a term known as polydispersity index (PDI). The average degree of polymerization ( DP ), which is not as important as the other three parameters, represents the average number of monomers that make up the polymer chains. N M N M i Mw 2 i i i i i i i i (Equation 2.1) i i N M N i Mn w M w i i (Equation 2.2) i i PDI DP Mw Mn Polymer molecular weight (M n ) Monomer molecular weight (Equation 2.3) (Equation 2.4) http://whale.wheelock.edu/bwcontaminants/analysis.htmL. 3 2. Experimental Procedure IMPORTANT!! Gloves MUST be worn when preparing the sample solution. 2.1 Sample Preparation WARNING: Anytime you are handling your sample or any solvents you MUST wear rubber gloves and have your hair tied back if it is long when preparing the sample. 1. Prepare your polymer sample in the following way: weigh out 25 mg of your polymer (i.e., four PS samples synthesized in Lab 1) with the balance and then add 5 mL of high performance liquid chromatography (HPLC) grade tetrahydrofuran (THF) so that the concentration is 5 mg/mL. 2. Filter the 5mL 5mg/mL PS THF solutions into clean vials. 2.2 Software Setup Check the status of UV lamp, it should be turned on. Push “shift+lamp” twice on the keyboard of UV detector to turn on the lamp. Make sure to turn off the UV lamp if you are last one to use GPC. 1. Double click on the Breeze icon on the desktop. The software window will look like that seen in Figure 2.2 below. 2. WARNING: DO NOT, AT ANY TIME, PRESS THE NOR THE (Purge Wizard) (Stop Pump Flow) BUTTONS. PRESSING THEM CAN CAUSE DAMAGE TO THE MACHINE. 4 Figure 2.2: Breeze Software. 3. Right click on the Pump Ramp Time button (it will look like this when depressed) and a window that will allow you to change the rate will pop up. Set the rate at 5 minutes and press OK. WARNING: Using a ramp time shorter than this will cause damage to the columns (e.g., it can change the packing of the polystyrene beads). 4. Move the Flow slider up until the flow rate is 1 mL/min then press the Go button below the Flow ruler. (If you do not press Go soon enough your rate will return to it previous setting.) 5. Press the Sample Queue button on the left. The spreadsheet seen in the middle right frame of Figure 1 will appear. Here the sample name and other parameters will have to be setup. 6. Type in a short sample name. In the Function column, click on the cell and select “Inject Broad Samples” from the dropdown menu. In the Method column, click on the cell and select “MatE 453_1” from the options. To set your Run Time, ask instructor how long it should. In the Injection Volume (Inj. Vol.) column, type in 5 a volume of “20”. In the Processing column (use the location bar at the bottom of the spread sheet to move to the right of spreadsheet where this column is located) select “Don’t process or report”. Step 5 and 6 can be eliminated, if only “single injection” is performed. 7. Next you need to set the baseline of your data graph to zero. When the system is ready, press the Equilibrate button. A popup window will appear asking you for the method of equilibration. Select “MatE 453_1” and press Equilibrate. In the real-time plot in the lower right hand corner of the window, a plot of the baseline can be monitored now. 8. On the detector output screen, monitor the set point. It should be zero; if it is not, press the “Auto zero” button. Keep pressing “Auto zero” until the set pint close to zero (when it stops fluctuating erratically). (If you press the Trace button on the key pad of the detector, you can see the same graph as in the real-time plot in the software window.) 9. Press the Abort Run button when the set point is zero. 2.3 Sample Injection 1. Under the chemical hood, pour about 25 mL of HPLC grade THF into a clean beaker. Use this to clean a calibrated syringe with a chromatography needle on it. NOTE: no sharp needles can be used with this system; they can cause the injector to leak. 2. Hold the syringe and the beaker with the same hand while using the other hand to pull the plunger in and out carefully yet quickly until all air bubbles are removed from the column. 3. Fill the syringe with about 25 l of your solution using the same method you used when cleaning the syringe. 4. Use the Make Single Injection icon to start the injection process. 5. A window will now appear asking for sample name, sample methods (select “MatE 453_1” from pull down window), sample type (select “broad unknown” from pull down window) and running time (type in 20 minutes). Hit inject button, a new window will pop up to instruct you on how to make the injection. Read 6 these directions and follow them. Take the red safety cap off the injection port and then the safety needle out of the injection hole. 6. Before injecting your sample into column, make sure your syringe tip is straight and the end of the tip is not sharp (should be flat). Stick the needle as far into the hole as possible (be sure you do not bend it while inserting). Inject the solution into the machine carefully so that the plunger is not bent. 7. Turn the black handle behind the injection port very quickly to the right (position 2). Wait five seconds and then turn it back to the original position 1. NOTE: You must turn the handle quickly so that the solution goes into the solution loop and not the solvent loop (see Figure 2.3). When the handle is in position 1, the solvent is delivered to the columns. When the handle is position 2, the solution is injected into the column. Figure 2.3 Manual Injector, back view. 8. The test will run for 25 minutes and the results will show on the right bottom corner in real time. Clean the syringe at least 4 times with THF as you did before. Put clear plastic needle cap back on the syringe needle and place in the box. Put the safety needle back into the injector port and replace the red safety cap. 9 When the data has been collected, you need to print your data. Click on the Find Data button in the left hand list. You will be given a list of the sample set you entered above. Double click on the name of the sample you want to print. A plot of your data will appear. Zoom in on one peak (ask your lab instructor which one if you have multiple peaks) by holding down the left mouse button and drawing a box around the peak of interest. 10 Select Print out of the File menu. In the Preferences select Landscape view. Then print the graph. Leave the machine on and DO NOT shut off the software. 3. Assignments 7 1). Based on the data obtained in the GPC experiment, and using a PS Standard calibration curve obtained previously (provided by the instructor), please perform the following calculations on three PS samples: (1) weight average molecular weight (Mw), (2) number average molecular weight (Mn), (3) polydispersity index (PDI) and (4) degree of polymerization ( DP ) (i.e., both Xw and Xn). Note: Please include at least one detailed calculation based on the GPC curve in your Lab Report. 2). Are there more than one peak in GPC curve? If so, explain why. 3). Based on the free radical polymerization kinetics, what is the expected order of molecular weight of four PS? And what is the experimental observation on the order of molecular weight of four PS? What causes the discrepancy between theory and experiment? 4). The wavelength of our UV detector is set as 254 nm. Why? Is this detector good for polyethylene samples? Please give a brief explanation. 8