Biochemical assessment - Springer Static Content Server
advertisement

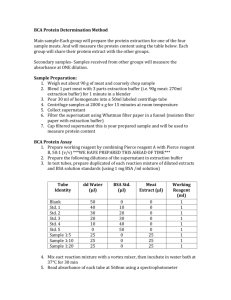
Biochemical assessment Biochemical tests were conducted immediately after last behavioral test. The animals were sacrificed by decapitation. Brains were removed and rinsed with ice-cold isotonic saline. Brains were then homogenized with ice-cold 0.1mmol/L phosphate buffer (pH 7.4). The homogenates (10%w/v) were then centrifuged at 10,000 g for 15 min and supernatants so formed were used for the biochemical estimations. Measurement of lipid peroxidation The extent of lipid peroxidation in the brain was determined quantitatively by performing the method as described by Wills [12]. The amount of malondialdehyde (MDA) was measured by reaction with thiobarbituric acid at 532 nm using Perkin Elmer Lambda 20 UV VIS Spectrophotometer (Norwalk, CT, USA). The values were calculated using the molar extinction co-efficient of chromophore (1.56 x 105 (mol/l)1 cm-1). Estimation of nitrite The accumulation of nitrite in the supernatant, an indicator of the production of nitric oxide was determined by a colorimetric assay with Greiss reagent (0.1 % N-(1napththyl) ethylene diamine dihydrochloride, 1% sulphanilamide and 5% phosphoric acid.) [4]. Equal volumes of the supernatant and Greiss reagent were mixed and incubated for 10 min at room temperature in the dark. The absorbance was measured at 540 nm using Perkin Elmer Lambda 20 UV VIS Spectrophotometer (Norwalk, CT, USA). The concentration of nitrite in the supernatant was determined from sodium nitrite standard curve. Estimation of reduced glutathione Reduced glutathione was estimated according to the method described by Ellman et al [1]. A 1 ml supernatant was precipitated with 1 ml of 4% sulphosalicylic acid and cold digested for 1 hour at 4°C. The samples were then centrifuged at 1,200 g for 15 min at 4°C. To 1 ml of the supernatant obtained, 2.7 ml of phosphate buffer (0.1mmol/l, pH 8) and 0.2 ml of 5, 5’ dithio-bis (2-nitrobenzoic acid) (DTNB) was added. The yellow color developed was measured at 412 nm using Perkin Elmer Lambda 20 UV VIS Spectrophotometer (Norwalk, CT, USA). Results were calculated using molar extinction co-efficient of the chromophore (1.36× 104 (mol/l)-1cm-1). Superoxide dismutase activity Superoxide dismutase (SOD) activity was assayed by the method of Kono [8]. The assay system consists of EDTA 0.1 mM, sodium carbonate 50 mM and 96 mM of nitro blue tetrazolium (NBT). In the cuvette, 2ml of the above mixture, 0.05 ml of hydroxylamine and 0.05 ml of the supernatant was added and auto-oxidation of hydroxylamine was measured for 2 min at 30s interval by measuring absorbance at 560 nm using Perkin Elmer Lambda 20 UV VIS Spectrophotometer (Norwalk, CT, USA). Catalase activity Catalase activity was assessed by the method of Luck [10], wherein the breakdown of H2O2 is measured. Briefly, assay mixture consists of 3 ml of H2O2 phosphate buffer and 0.05 ml of the supernatant of the tissue homogenate. The change in absorbance was recorded for 2 min at 30 s interval at 240 nm using Perkin Elmer Lambda 20 spectrophotometer. The results are expressed as micromoles of hydrogen peroxide decomposed per min per mg of protein. Glutathione-S-transferase activity The activity of glutathione–s-transferase was assayed by the method of Habig and Jakoby [5]. Briefly, the assay mixture consisted of 2.7 ml of phosphate buffer, 0.1 ml of reduced glutathione, 0.1 ml of 1-chloro-2, 4-dinitrobenzene (CDNB) as substrate and 0.1 ml of supernatant. The increase in the absorbance was recorded at 340 nm for 5 min at 1 min interval using Perkin Elmer Lambda 20 UV VIS Spectrophotometer (Norwalk, CT, USA). The results are expressed as nmoles of CDNB conjugated/min/mg of protein. Estimation of Acetyl cholinesterase (AChE) activity AchE is a marker of loss of cholinergic neurons in the forebrain. The AChE activity was assessed by Ellman method [2]. The assay mixture contained 0.05 ml of supernatant, 3 ml of sodium phosphate buffer (pH 8), 0.1 ml of acetylthiocholine iodide and 0.1 ml of DTNB (Ellman reagent). The change in absorbance was measured for 2 min at 30s interval at 412 nm using Perkin Elmer Lambda 20 UV VIS Spectrophotometer (Norwalk, CT, USA). Results are expressed as micromoles of acetylthiocholine iodide hydrolyzed per min per mg of protein. Protein estimation The protein content was estimated by Biuret method [3] using bovine serum albumin as a standard Aluminum estimation The aluminium was analyzed by wet acid digestion method of Zumkley [13] in hippocampus and cortex of the brain. A mixture of 2.5 ml of per choleric acid/nitric acid (1:4 in volume) was added to brain part which is then placed in sand bath for 44 h until the point of a white ash or residue obtained. Residues were dissolved in 2.5 ml of 10mM nitric acid. Then this sample (in liquid form) was placed in the sample holder of atomic absorption spectrophotometer (Perkin Elmer, India). The total concentration of aluminium was calculated in μg/gm of tissue. Mitochondrial Complex Estimation Isolation of rat brain mitochondria The whole brain (excluding cerebellum) was used for mitochondrial isolation. Rat brain mitochondria were isolated by differential centrifugation. The brain is homogenized in 10 ml of homogenizing buffer containing 225 mM mannitol,75 mM sucrose, 5 mM HEPES, 1mM EGTA, 1 mg/ml BSA, pH 7.4. The homogenate is brought to 30 ml with the same buffer and centrifuged at 2000 g for 3 min at 40C. The pellet is discarded and the supernatant is divided into 2 tubes and centrifuged at 12000g for 10 min. The pellet containing the mixture of synaptosomes and mitochondria is suspended in 10 ml of homogenization buffer containing 0.02% digitonin to lyse the synaptosomes followed by centrifugation at 12000g for 10 min to pellet down both extra synaptosomal and intra synaptosomal mitochondria. The mitochondrial pellet is washed twice in the same buffer without EGTA, BSA and digitonin. Complex-I (NADH Dehydrogenase activity) Complex-I was measured spectrophotometrically by the method of King and Howard [7]. The method involves catalytic oxidation of NADH to NAD+ with subsequent reduction of cytochrome C. The reaction mixture contained 0.2 M glycyl glycine buffer pH 8.5, 6 mM NADH in 2 mM glycyl Glycine buffer and 10.5 mM cytochrome C. The reaction was initiated by addition of requisite amount of solubilized mitochondrial sample and followed absorbance change at 550 nm for 2 min. Complex-II (Succinate Dehydrogenase (SDH) activity) SDH was measured spectrophotometrically according to King [6]. The method involves oxidation of succinate by an artificial electron acceptor, potassium ferricyanide. The reaction mixture contained 0.2 M phosphate buffer pH 7.8, 1% BSA, 0.6 M succinic acid, and 0.03 M potassium ferricyanide. The reaction was initiated by the addition of mitochondrial sample and absorbance change was followed at 420 nm for 2 min. Complex-III (MTT ability) The MTT assay is based on the reduction of (3-(4, 5- dimethylthiazol-2-yl)-2, 5diphenyl-H-tetrazolium bromide (MTT) by hydrogenase activity in functionally intact mitochondria. The MTT reduction rate was used to assess the activity of the mitochondrial respiratory chain in isolated mitochondria by the method of Liu et al. [9]. Briefly, 100 μl mitochondrial samples were incubated with 10 μl MTT for 3 hour at 37 °C. The blue formazan crystals were solubilized with dimethylsulfoxide and measured by an ELISA reader at 580 nm filter Complex-IV (Cytochrome oxidase assay) Cytochrome oxidase activity was assayed in brain mitochondria according to the method of Sotocassa [11]. The assay mixture contained 0.3 mM reduced cytochrome C in 75 mM phosphate buffer. The reaction was started by the addition of solubilized mitochondrial sample and absorbance change was recorded at 550 nm for 2 min. Reference 1. Ellman GL (1959) Tissue Sulfhydryl Groups. Archives of Biochemistry and Biophysics 82:70-77. 2. Ellman GL, Courtney KD, Andres V, Featherstone RM (1961) A New and Rapid Colorimetric Determination of Acetylcholinesterase Activity. Biochemical Pharmacology 7:88-&. 3. Gornall AG, Bardawill CJ, David MM (1949) Determination of Serum Proteins by Means of the Biuret Reaction. Journal of Biological Chemistry 177:751-766. 4. Green LC, Wagner DA, Glogowski J, Skipper PL, Wishnok JS, Tannenbaum SR (1982) Analysis of Nitrate, Nitrite, and [N-15]-Labeled Nitrate in Biological-Fluids. Analytical Biochemistry 126:131-138. 5. Habig WH, Jakoby WB (1981) Assays for differentiation of glutathione Stransferases. Methods Enzymol 77:398-405. 6. King TE (1967) [58] Preparation of succinate dehydrogenase and reconstitution of succinate oxidase. In: Methods in Enzymology, vol. Volume 10 (Ronald W. Estabrook, M. E. P., ed), pp 322-331: Academic Press. 7. King TE, Howard RL (1967) [52] Preparations and properties of soluble NADH dehydrogenases from cardiac muscle. In: Methods in Enzymology, vol. Volume 10 (Ronald W. Estabrook, M. E. P., ed), pp 275-294: Academic Press. 8. Kono Y (1978) Generation of Superoxide Radical during Autoxidation of Hydroxylamine and an Assay for Superoxide-Dismutase. Archives of Biochemistry and Biophysics 186:189-195. 9. Liu Y, Peterson DA, Kimura H, Schubert D (1997) Mechanism of cellular 3(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction. J Neurochem 69:581-593. 10. Luck H (1971) Catalase. Methods of Enzymatic analysis Bergmeyer HU (Eds). Academic Press, New York. 885-893. 11. Sottocasa GL, Kuylenstierna B, Ernster L, Bergstrand A (1967) An electrontransport system associated with the outer membrane of liver mitochondria. A biochemical and morphological study. The Journal of cell biology 32:415-438. 12. Wills ED (1966) Mechanisms of lipid peroxide formation in animal tissues. Biochem J 99:667-676. 13. Zumkley H, Bertram HP, Lison A, Knoll O, Losse H (1979) Aluminum, zinc and copper concentrations in plasma in chronic renal insufficiency. Clin Nephrol 12:18-21.