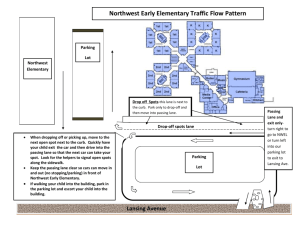
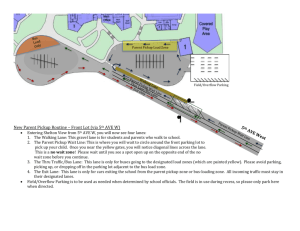
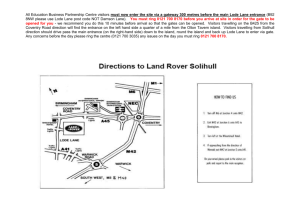
srep01121-s1
advertisement

SUPPLEMENTARY DATA Equilibrious Strand Exchange Promoted by DNA Conformational Switching Zhiguo Wu1, Xiao Xie1, Puzhen Li1, Jiayi Zhao1, Lili Huang1 and Xiang Zhou*1, 2 1 College of Chemistry and Molecular Sciences, Key Laboratory of Biomedical Polymers of Ministry of Education, Wuhan University, Wuhan, Hubei, 430072, P. R. of China. 2 State Key Laboratory of Natural and Biomimetic Drugs, Peking University, Beijing, P. R. of China. Table S1. The DNA sequences with and without c-myc (mu-c-myc). Primers Sequence (5’-3’) label GCATCTACGTATCGTAGATGCATT 24 bp free AATGCATCTACGATACGTAGATGC 24 nt AATGCATCTACGATACGTAGATGC 5’-Cy5 TGGGGAGGGTGGGGAGGGTGGGGT c-myc 24 bp free ACCCCACCCTCCCCACCCTCCCCA c-myc 24 nt G4-24 nt ACCCCACCCTCCCCACCCTCCCCA 5’-Cy5 TGGGGAGGGTGGGGAGGGTGGGGT 5’-Cy5 TGCTTACGCTGTGCATCTACGTATCGTAGATGCATTTGAGGAT AGGCT 48 bp free AGCCTATCCTCAAATGCATCTACGATACGTAGATGCACAGCGT AAGCA 48 nt AGCCTATCCTCAAATGCATCTACGATACGTAGATGCACAGCGT AAGCA 5’-Cy5 TGCTTACGCTGTTGGGGAGGGTGGGGAGGGTGGGGTTGAGG ATAGGCT c-myc 48 bp free AGCCTATCCTCAACCCCACCCTCCCCACCCTCCCCAACAGCGT AAGCA c-myc 48 nt AGCCTATCCTCAACCCCACCCTCCCCACCCTCCCCAACAGCGT AAGCA 5’-Cy5 G4-48 nt TGCTTACGCTGTTGGGGAGGGTGGGGAGGGTGGGGTTGAGG ATAGGCT 5’-Cy5 ACGTATCTACTTTGCTTACGCTGTGCATCTACGTATCGTAGATG CATTTGAGGATAGGCTCGTTCAACTGTA 72 bp free TACAGTTGAACGAGCCTATCCTCAAATGCATCTACGATACGTA GATGCACAGCGTAAGCAAAGTAGATACGT 72 nt TACAGTTGAACGAGCCTATCCTCAAATGCATCTACGATACGTA GATGCACAGCGTAAGCAAAGTAGATACGT 5’-Cy5 ACGTATCTACTTTGCTTACGCTGTTGGGGAGGGTGGGGAGGG TGGGGTTGAGGATAGGCTCGTTCAACTGTA c-myc 72 bp free TACAGTTGAACGAGCCTATCCTCAACCCCACCCTCCCCACCCT CCCCAACAGCGTAAGCAAAGTAGATACGT c-myc 72 nt G4-72 nt TACAGTTGAACGAGCCTATCCTCAACCCCACCCTCCCCACCCT CCCCAACAGCGTAAGCAAAGTAGATACGT ACGTATCTACTTTGCTTACGCTGTTGGGGAGGGTGGGGAGGG TGGGGTTGAGGATAGGCTCGTTCAACTGTA 5’-Cy5 5’-Cy5 TGGGGAGGGTGGAGAGGGTGGGGT mu-c-myc 24bp free ACCCCACCCTCTCCACCCTCCCCA mu-c-myc 24 nt ACCCCACCCTCTCCACCCTCCCCA 5’-Cy5 TGCTTACGCTGTTGGGGAGGGTGGAGAGGGTGGGGTTGAGG ATAGGCT mu-c-myc 48 bp free AGCCTATCCTCAACCCCACCCTCTCCACCCTCCCCAACAGCGT AAGCA mu-c-myc 48 nt AGCCTATCCTCAACCCCACCCTCTCCACCCTCCCCAACAGCGT AAGCA 5’-Cy5 ACGTATCTACTTTGCTTACGCTGTTGGGGAGGGTGGAGAGGG TGGGGTTGAGGATAGGCTCGTTCAACTGTA mu-c-myc 72 bp free TACAGTTGAACGAGCCTATCCTCAACCCCACCCTCTCCACCCT CCCCAACAGCGTAAGCAAAGTAGATACGT mu-c-myc 72 nt TACAGTTGAACGAGCCTATCCTCAACCCCACCCTCTCCACCCT 5’-Cy5 CCCCAACAGCGTAAGCAAAGTAGATACGT ACGTATCTACTTTGGGGAGGGTGGGGAGGGTGGGGTTGGGG AGGGTGGGGAGGGTGGGGTCGTTCAACTGTA 2c-myc 72 bp free TACAGTTGAACGACCCCACCCTCCCCACCCTCCCCAACCCCAC CCTCCCCACCCTCCCCAAAGTAGATACGT 2c-myc 72 nt TACAGTTGAACGACCCCACCCTCCCCACCCTCCCCAACCCCAC CCTCCCCACCCTCCCCAAAGTAGATACGT 5’-Cy5 Figure S1. The PAGE analysis of the SERs of dsDNAs and homologous ssDNAs. TE buffer was used instead of sodium phosphate buffer. The invading strands contained the complementary strand of c-myc. Lane 1: SERs of random sequences; lane 2: SERs of c-myc containing sequences; lane 3: dsDNAs markers of random sequences; lane 4: dsDNAs markers of c-myc sequences; lane 5: ssDNAs markers of random sequences; lane 6: ssDNAs markers of c-myc sequences. The SERs were carried out in the presence of 200 mM KCl, 20 mM TE, pH 7.4. The incubation temperature was 37 oC, and the incubation times of 24 bp and 24 nt, 48 bp and 48 nt, 72 bp and 72 nt, 2c-myc 72 bp and 72nt were 3 h, 30 h, 60 h, 60 h, respectively. Figure S2. The CD spectrum of c-myc and mu-c-myc sequences (single strands) at the condition of: 200 mM KCl, 20 mM sodium phosphate, pH 7.4. 5 μM ssDNA for c-myc and mu-c-myc. Figure S3. The CD spectra of the duplexes containing c-myc (were in red, c-myc 24 bp, A; c-myc 48 bp, B; c-myc 72 bp, C) and not containing c-myc (were in black, random 24 bp, A; random 48 bp, B; random 72 bp, C). In the right panel, the CD spectra of a, c-myc 24 bp – random 24 bp; b, c-myc 48 bp – random 48 bp; c, c-myc 72 bp – random 72 bp were obtained. The CD spectra were obtained at the same conditions to the SERs (200 mM KCl, 20 mM sodium phosphate, pH 7.4). The dsDNAs concentrations were 5 μM, 2 μM and 2 μM for 24 bp, 48 bp and 72 bp, respectively. Figure S4. In left panel, the CD spectra of mu-c-myc 24 bp and random 24 bp, mu-c-myc 48 bp and random 48 bp, mu-c-myc 72 bp and random 72 bp were recorded. In right panel, the difference values of mu-c-myc sequences and random sequences were obtained. The CD spectra were performed at 5 μM, 2 μM and 2 μM for 24 bp, 48 bp and 72 bp, respectively, under the condition of 200 mM KCl, 20 mM sodium phosphate, pH 7.4. Figure S5. The c-myc promoted SER kenetics. The SERs DNAs were adjusted to 20 μM dsDNAs (bp) and 20 μM ssDNAs (nt), buffered by 200 mM KCl, 20 mM sodium phosphate, pH 7.4. Then the SERs were carried out by gradiently increasing the incubating time from 0 to 7 h, 33 h and 72 h for A, B, and C, respectively. The SERs were stopped by cooling to 4 oC, and then bromophenol blue was added as the loading buffer. 20% PAGE gel containing 200 mM KCl, 20 mM TBE (pH 7.4) was employed to electrophorese the DNA samples. All the c-myc contained sequences (green) and random sequences (yellow) were placed together with the PAGE gel results. The Cy5-labeled complementary strands of c-myc contained sequences were red, and the Cy5-labeled complementary strands of random sequences were yellow. Figure S6. The Tm values of the random sequence ssDNAs. The normalized Tm experiments were carried out at 10 μM, 5 μM, 2 μM single strand for 24 nt and 24 nt com, 48 nt and 48 nt com, 72 nt and 72 nt com, respectively. In the same buffer as SERs, 200 mM KCl, 20 mM sodium phosphate, pH 7.4. The system temperatures were increased gradually from 25 oC to 90 oC, and the ultraviolet absorbance at 260 nm were monitored to calculate the Tm values for each ssDNA. Figure S7. The denaturing PAGE gel electrophoresis of Cy5 labeled ssDNAs. 6 M urea and 40% formamide were added into 20% PAGE gel to denature the single stranded DNAs. The experiment was carried out in the buffer containing 20 mM KCl, 20 mM TBE, pH 7.4. Lane 1: random 24 nt com; lane 2: c-myc 24 nt com; lane 3: random 48 nt com; lane 4: c-myc 48 nt com; lane 5: random 72 nt com; lane 6: c-myc 72 nt com. The concentration of ssDNAs was 2μM. Figure S8. The Tm values of the dsDNAs with or without c-myc. The normalized Tm experiments were carried out at 5 μM, 2 μM, 2 μM double strand for 24 bp (A), 48 bp (B), 72 bp (C) lengths DNA, respectively. In the same buffer as SERs, 200 mM KCl, 20 mM sodium phosphate, pH 7.4. The system temperatures were increased gradually from 25 oC to 90 oC, and the ultraviolet absorbances at 260 nm were monitored to calculate the Tm values for each duplex. Figure S9. The UV-melting temperature of all the double stranded DNAs used in this paper, recorded at 260 nm. The Tm experiments were performed 5 μM, 2 μM and 2 μM for 24 bp, 48 bp and 72 bp, respectively, under the condition of 20 mM KCl, 20 mM NaCl, 20 mM TE, pH 7.4. Figure S10. The SER of 2c-myc sequence as the control. A, the PAGE analysis of the SER, lane 1: SER of random 72 bp and 72 nt; lane 2: SER of c-myc 72 bp and 72 nt; lane 3: SER of 2c-myc 72 bp and 72 nt; lane 4: dsDAN marker of random sequence; lane 5: dsDNA marker of c-myc sequence; lane 6: dsDNA marker of 2c-myc sequence; lane 7: ssDNA marker of random sequence; lane 8: ssDNA marker of c-myc sequence; lane 9: ssDNA marker of 2c-myc sequence. These SERs were carried out at 37 oC, 60 h, in the buffer of 200 mM KCl, 20 mM sodium phosphate, pH 7.4. B, the histogram of SERs product concentrations extracted from A, the random sequence, c-myc sequence and 2c-myc sequence. Figure S11. The SERs of dsDNAs and ssDNAs with mutated c-myc (mu-c-myc) and with the random sequences. Lane 1: the random sequences; lane 2: the mu-c-myc contained sequences; lane 3: dsDNAs with random sequence; lane 4: dsDNAs with the mu-c-myc sequence; lane 5: ssDNAs with random sequence; lane 6: ssDNAs with mu-c-myc sequence. The SERs were carried out under the condition of 200 mM KCl, 20 mM sodium phosphate, pH 7.4. The incubation times of 24 bp and 24 nt, 48 bp and 48 nt, 72 bp and 72 nt were 3 h, 30 h, 60 h, respectively. And the incubation was performed at 37 oC. Figure S12. The SERs containing single-base mismatch system. Lane 1: random sequences without c-myc; lane 2: c-myc contained sequences; lane 3: the sequences containing 1 mismatch in the double strands, and the Cy5-labeled sequences were the fully complementary strands of the mu-c-myc contained strands; lane 4: dsDNAs with random sequence; lane 5: dsDNAs with mu-c-myc sequence; lane 6: ssDNAs with random sequence; lane 7: ssDNAs with mu-c-myc sequence. The SERs were performed under the same conditions to Figure 2 in main text. The single-base mismatch sequences were presented under the PAGE gel. Figure S13. The random sequences and c-myc containing sequences formed double stranded DNA duplexes under the SERs conditions. Lane 1: DNA markers; lane 2: random 24 bp; lane 3: c-myc 24 bp; lane 4: random 48 bp; lane 5: c-myc 48 bp; lane 6: random 72 bp; lane 7: c-myc 72 bp; lane 8: 2c-myc 72 bp. The DNAs were incubated at 37 oC, 200 mM KCl, 20 mM sodium phosphate, pH 7.4. Then native PAGE was employed to analysis the DNA samples, stained by EB, and the image was collected by UV imager. Figure S14. The SERs between dsDNAs and Cy5-labeled ssDNAs mediated by Rec A in the absence of ATP. Lane 1: random 24 bp, nt; lane 2: c-myc 24 bp, nt; lane 3: random 48 bp, nt; lane 4: c-myc 48 bp, nt; lane 5: random 72 bp, nt; lane 6: c-myc 72 bp, nt. All the SER conditions were identical with the experiment in Figure 6 except ATP, and no SER product was observed. Figure S15. The Tm values of the double stranded DNAs in the presence of 40% PEG200. The normalized Tm experiments were carried out at 5 μM, 2 μM, 2 μM dsDNAs for 24 bp and c-myc 24 bp, 48 bp and c-myc 48 bp, 72 bp and c-myc 72 bp, respectively. The buffer contains 5 mM KCl, 5 mM TE, and 40% PEG200 as the crowding agent pH 7.4. The system temperatures were increased gradually from 25 oC to 90 oC, and the ultraviolet absorbance at 260 nm were monitored to calculate the Tm values for each dsDNA.