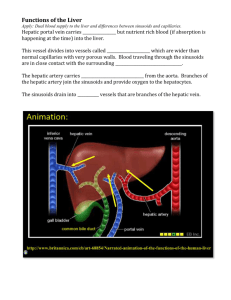
Protocol for hepatocytes and hepatic stellate cells Isolation, Culture
advertisement

Name & Project (beachte: modifizierter Titel) – B2 Steven Dooley - Impact of Stellate Cell-Hepatocyte Communication to (TGF-) Signal Transduction and Cell Fate during Liver Damage, Inflammation and Regeneration - Identification of Relevant Cytokines - Modulation of Signaling Interpretation - Link to Organ Level Interaction (partial hepatectomy) State of the art, and own contributions Hepatic stellate cells (HSCs), Kupffer cells and immune cells converge in a complex interaction with hepatocytes to provoke inflammatory and/or regenerative responses in the damaged liver. Uncovering the specific effects of growth factors on these cells, defining the interaction of different cell populations mediating these responses will enable the discovery of new therapeutic approaches. HSCs are resident vitamin A-storing cells in the perisinusoidal space of Disse between the sinusoidal endothelium and hepatocytes. Following hepatic injury, HSCs become activated, proliferate and produce extracellular matrix (ECM) (Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology 2008; 134:1655–1669). Both the characterization of HSCs and their behavior in hepatic injury have been well characterized, and excellent models exist for their analysis in vivo and in culture (Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev 2008; 88:125–172). Transforming growth factor (TGF-) and platelet derived growth factor (PDGF) are considered the key stimuli for HSC activation and proliferation, respectively. Members of the TGF-β cytokine family exist in variant forms (e.g., TGF-β1, 2 and 3; bone morphogenic proteins (BMPs)). TGF-β signals via serine/threonine kinase receptors. TGF-β binding to type II receptors elicits a response where type I receptors are phosphorylated and activated. The latter then propagates the signal by phosphorylating Smad transcription factors, the so-called receptor Smads (R-Smads). Receptors of the TGF-β cytokine family phosphorylate Smads 2 and 3, whereas BMP receptors phosphorylate Smads 1, 5, and 8. Once activated, the R-Smads complex Smad4, a binding partner common to all R-Smads, and translocate to the nuclear compartment (Shi Y, Massague J: Mechanisms of tgf-beta signaling from cell membrane to the nucleus. Cell 2003;113:685-700). In order to achieve binding with high affinity and selectivity to specific target genes the Smad4-R-Smad complexes associate with additional DNA-binding cofactors (Massague J, Seoane J, Wotton D: Smad transcription factors. Genes Dev 2005;19:2783-2810). Each Smad4/R-Smad-cofactor combination targets a particular set of genes. Consecutively, a TGF-β stimulus can activate or repress several hundreds of target genes at the same time by virtue of combinatorial interactions (Massague J: Tgfbeta in cancer. Cell 2008;134:215-230). In healthy human livers TGF-1 transcripts and protein expression are absent. After partial hepatectomy, TGF- transcripts are detectable in all cell types (Bissell DM, Wang SS, Jarnagin WR, Roll FJ: Cell-specific expression of transforming growth factor-beta in rat liver. Evidence for autocrine regulation of hepatocyte proliferation. J Clin Invest 1995;96:447-455). TGF-β signaling is negatively regulated by inhibitory Smads, such as Smad7 and Smad6 (Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, Topper JN, Gimbrone MA, Jr., Wrana JL, Falb D: The mad-related protein smad7 associates with the tgfbeta receptor and functions as an antagonist of tgfbeta signaling. Cell 1997;89:1165-1173; Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata N, Heldin NE, Heldin CH, Tendijke P: Identification of smad7, a tgf beta-inducible antagonist of tgf-beta signalling. Nature 1997;389:631-635). TGF-β not only enhances the expression of Smad7 transcription but also mobilizes a pre-existing nuclear pool of Smad7 to inhibit TGF-β receptors (Itoh S, Landstrom M, Hermansson A, Itoh F, Heldin CH, Heldin NE, Tendijke P: Transforming growth factor beta 1 induces nuclear export of inhibitory smad7. J Biol Chem 1998;273:2919529201). Smad7 expression can also be induced by other signaling inputs, e.g., IFN-, TNF- and EGF (Park SH: Fine tuning and cross-talking of tgf-beta signal by inhibitory smads. J Biochem Mol Biol 2005;38:9-16). Smad7 represses TGF-β signaling by stably binding to the cytoplasmic domain of activated type I receptors and thereby competing with R-Smads for receptor activation. Since 1999 our group has been working on TGF- signaling in liver cells. At approximately that time, Smad7 was identified as a negative regulatory protein in the TGF- signalling pathway. We have delineated the genomic structure of the rat Smad7 gene and discovered that it is transiently induced by TGF- in quiescent HSCs (Stopa M, Benes V, Ansorge W, Gressner AM, Dooley S: Genomic locus and promoter region of rat smad7, an important antagonist of tgfbeta signaling Mamm Genome 2000;11:169-176). We have dissected a TGF- response element within the Smad7 promoter and identified a TGF--induced stimulatory binding complex, comprising of Smad2, Smad3 and Smad4 (Stopa M, Anhuf D, Terstegen L, Gatsios P, Gressner AM, Dooley S: Participation of smad2, smad3, and smad4 in transforming growth factor beta (tgf-beta)-induced activation of smad7. The tgf-beta response element of the promoter requires functional smad binding element and e- box sequences for transcriptional regulation. J Biol Chem 2000;275:29308-29317). By analyzing the activation process of HSCs, it became clear that TGF signaling is modulated when comparing quiescent and activated HSCs (Dooley S, Delvoux B, Lahme B, Mangasser-Stephan K, Gressner AM: Modulation of transforming growth factor beta response and signaling during transdifferentiation of rat hepatic stellate cells to myofibroblasts. Hepatology 2000;31:1094-1106; Dooley S, Delvoux B, Streckert M, Bonzel L, Stopa M, ten Dijke P, Gressner AM: Transforming growth factor beta signal transduction in hepatic stellate cells via smad2/3 phosphorylation, a pathway that is abrogated during in vitro progression to myofibroblasts. Tgfbeta signal transduction during transdifferentiation of hepatic stellate cells. FEBS Lett 2001;502:4-10; Dooley S, Streckert M, Delvoux B, Gressner AM: Expression of smads during in vitro transdifferentiation of hepatic stellate cells to myofibroblasts. Biochem Biophys Res Commun 2001;283:554-562). Among others, activated HSCs do not respond to TGF- with an induction of Smad7 expression, obviously one premise for the chronicity of TGF activities during disease progression. Our hypothesis was further supported by a gene therapeutic approach. Smad7 overexpression in damaged rat livers blunted Smad C-terminal phosphorylation, arrested rat HSCs in a quiescent stage and reduced liver damage, indicating the predominant role of TGF- in HSC activation and suggesting an antifibrotic potential for Smad7 (Dooley S, Hamzavi J, Breitkopf K, Wiercinska E, Said HM, Lorenzen J, Ten Dijke P, Gressner AM: Smad7 prevents activation of hepatic stellate cells and liver fibrosis in rats. Gastroenterology 2003;125:178-191). At that time we assumed that antifibrotic effects of Smad7 were directed towards HSCs, since TGF- signaling in hepatocytes was believed to result in proliferation arrest and apoptosis. However, when quantifying effects on hepatocytes in injured livers of mouse models and human patients, TGF- signaling was present and excelled the occurrence of apoptosis, indicating further functions of TGF- in this cell type. Analysing TGF- expression signatures over time, EMT-related gene regulation (Dooley S, Hamzavi J, Ciuclan L, Godoy P, Ilkavets I, Ehnert S, Ueberham E, Gebhardt R, Kanzler S, Geier A, Breitkopf K, Weng H, Mertens PR: Hepatocyte-specific smad7 expression attenuates tgf-beta-mediated fibrogenesis and protects against liver damage. Gastroenterology 2008;135:642-659; Meindl-Beinker NM, Dooley S: Transforming growth factor-beta and hepatocyte transdifferentiation in liver fibrogenesis. J Gastroenterol Hepatol 2008;23 Suppl 1:S122-127; Weng HL, Ciuclan L, Liu Y, Hamzavi J, Godoy P, Gaitantzi H, Kanzler S, Heuchel R, Ueberham U, Gebhardt R, Breitkopf K, Dooley S: Profibrogenic transforming growth factor-beta/activin receptor-like kinase 5 signaling via connective tissue growth factor expression in hepatocytes. Hepatology 2007;46:1257-1270) occurred in monolayer cultured mouse hepatocytes and was reversed by interference with TGF- signaling. Furthermore, spontaneous transdifferentiation of hepatocytes was reversible, when transferring monolayer cells into a three-dimensional “sandwich” culture system. This provides an ECM (micro)environment required for maintenance of polarized epithelial morphology (Godoy P, Hengstler JG, Ilkavets I, Meyer C, Bachmann A, Muller A, Tuschl G, Mueller SO, Dooley S: Extracellular matrix modulates sensitivity of hepatocytes to fibroblastoid dedifferentiation and transforming growth factor beta-induced apoptosis. Hepatology 2009). Thus, TGF- orchestrates a complex network of inter- and intracellular communications, altogether directing the (patho)physiological response of the organ. Amongst others, HSC activation, extracelluar matrix (ECM) synthesis, induction of important pro-fibrogenic and tumorigenic mediators (e.g. CTGF, TIMP-1, Col1, PAI-1, PDGF) and epithelial to mesenchymal transition of hepatocytes and other cell types of the liver may be summarized under the TGF-dependent effects. TGF- very actively participates in the process of liver regeneration, especially in terminating hepatocyte proliferation after partial hepatectomy (Michalopoulos GK: Liver regeneration. J Cell Physiol 2007;213:286-300) and a massive deregulation may lead in its worst scenario to liver cancer and metastasis. To further delineate TGF--dependent mechanisms, a transgenic mouse model with inducible and hepatocyte-specific Smad7 expression able to interfere with TGF- signaling in this cell type was generated. Switching on the C-reactive protein promoter with lipopolysaccharide in these animals results in hepatocyte-specific expression of Smad7. Testing these mice in a chronic model of liver damage (CCl4 treatment for 8 weeks) permits LPS/Smad7-dependent abrogation of TGF- signaling and improves liver damage (Dooley S, Hamzavi J, Ciuclan L, Godoy P, Ilkavets I, Ehnert S, Ueberham E, Gebhardt R, Kanzler S, Geier A, Breitkopf K, Weng H, Mertens PR: Hepatocyte-specific smad7 expression attenuates tgf-beta-mediated fibrogenesis and protects against liver damage. Gastroenterology 2008;135:642-659). As a complementary animal model, we used a mouse strain with a disrupted Exon1 at the Smad7 genomic locus, S7E1, thus lacking a fully functional Smad7 gene. In contrast to the Smad7 overexpressing model, these mice display enhanced liver damage and fibrogenesis upon challenge with CCl4 (Hamzavi J, Ehnert S, Godoy P, Ciuclan L, Weng H, Mertens PR, Heuchel R, Dooley S: Disruption of the smad7 gene enhances cci4-dependent liver damage and fibrogenesis in mice. J Cell Mol Med 2008;12:21302144). These results suggest that a functional TGF- signaling pathway in hepatocytes is required for disease progression in the chronically injured liver and further underlines a beneficial impact of the TGF- antagonist Smad7, at least at the early stages of chronic liver disease. On the other hand, when performing time course analyses with liver tissue in BDL rats, a continously increasing Smad7 expression becomes apparent that correlates with severity of liver damage (Seyhan H, Hamzavi J, Wiercinska E, Gressner AM, Mertens PR, Kopp J, Horch RE, Breitkopf K, Dooley S: Liver fibrogenesis due to cholestasis is associated with increased smad7 expression and smad3 signaling. J Cell Mol Med 2006;10:922-932). Description of planned work for five years, including clearly formulated milestones after three years (maximum 1.5 page) Cell preparation Mouse hepatocytes will be isolated according to the established SOP, resulting in 95% purity and a viability of 85%, as determined by trypan blue exclusion. Mouse hepatic stellate cells (HSCs) will be prepared by a two-step protocol (collagenase/pronase digestion and Nycodenz gradient). Because of their lipid content, stellate cells are the least dense fraction of the nonparenchymal cells, and during centrifugation they float effectively away from other hepatic cells. HSCs are identified by their characteristic lipid droplet inclusion (light microscopy, autofluorescence) and positive vimentin staining. Purity is usually between 90% to 95%, vitality 80% as estimated by Trypan blue exclusion test. Kupffer cells were not detected (phagocytosis of fluorescent latex beads, ED2 antibody staining). Cells isolated from 3 mice (approximately 5 x 106 cells per mouse) are seeded on one 60-mm uncoated plastic tissue culture dish and cultured in DMEM supplemented with 10% fetal calf serum (FCS) and standard antibiotics in 95% air 5% CO2 humidified atmosphere at 37°C. HSCs are termed quiescent until day 2 of culture and activated (myofibroblasts) at day 7. Culture days 3, 4 and 5 represent intermediate activation stages. Cultured MFBs can be passaged after 5 to 7 days in culture and represent fully activated MFBs after reseeding. Purified cells can be frozen as previously described (Cryopreservation of hepatic stellate cells. Neyzen S, Van de Leur E, Borkham-Kamphorst E, Herrmann J, Hollweg G, Gressner AM, Weiskirchen R. J Hepatol. 2006 May;44(5):910-7) and shipped with Fedex to other groups within the network. One aim of this project is establishing an SOP for the simultaneous isolation of stellate cells and hepatocytes from the same mouse liver tissue, as it was described for rat liver (Riccalton-Banks L, Bhandari R, Fry J, Shakesheff KM. Mol Cell Biochem. 2003 Jun;248(12):97-102e). Although it is not critical to use cells from different animals of the same strain, to reduce workload and save animals, it is intended to prepare HSCs and hepatocytes simultaneously. HSCs and hepatocyte communication Conditioned media from mouse HSCs and hepatocytes will be collected at different time points (culture days 2 to 7) and frozen at -80oC until use. These media will be used to study the impact of HSCs towards hepatocytes and vice versa with emphasis on TGF- signal transduction. Alternatively, two different HSCs-hepatocytes coculture systems will be setup for this purpose, as previously described (Uyama N, Shimahara Y, Kawada N, Seki S, Okuyama H, Iimuro Y, Yamaoka Y, Regulation of cultured rat hepatocyte proliferation by stellate cells. J Hepatol. 2002, 36:590-9). One without direct contact of HSCs and hepatocytes for biochemical analyses, where it is required to work off the cells separately and one with direct cell cell contact, which will be useful for imaging purposes/microscopy at the single cell level, where the different cell types can be discriminated by eye. Generation of experimental data Cell culture Molecular mechanisms regulating TGF-β mediated signalling in HSCs are examined by data based mathematical modelling, exactly as previously done for hepatocytes, including R-Smad2/3 activation and trafficking as well as negative feedback regulation via SnoN and Smad7 (the latter is expected to be of high relevance in HSCs). Modulation of signaling by HSC-hepatocyte communication will be integrated. Further, these studies will be extended to include Smad1 dependent TGF-β signalling. Time resolved genome wide expression profiling will be done to identify influences of HSCs and hepatocytes to eachother, with special impact for TGF-β signatures. The absolute number of molecules per cell will be determined, as previously done in cultured hepatocytes, for relevant pathway components as listed. In addition, quantitative proteomics will be done with the successful nonlabelling quantification method based on LCMALDIMS. Herewith, it will be possible to determine, e.g. the SMAD2/SMAD3 concentration in good accordance with quantitative immunoblotting experiments from the Klingmüller group. Furthermore, customized peptide arrays, like reverse phase protein microarrays (RPPA) and microspot immunoassays (MIA), as described in subproject D3 will be applied to produce quantitative biochemical data. Thus, a dynamic pathway model of TGF--Smad signal transduction in HSCs will be established accordingly based on multiple quantitative immunoblotting and ELISA data sets. Moreover, model-derived predictions for TGF-- signaling in hepatocytes, e.g. the perturbations of SnoN-mediated negative feedback regulation, which were experimentally validated, have now to be proven within the context of a HSC-hepatocyte communication, measuring the impact of conditioned media and in coculture of both. An siRNA-mediated knockdown approach, which was successfully established for cultured human and mouse hepatocytes, will be developed for HSCs. Animal experiments Mice will be challenged either with partial hepatectomy or a single application of the liver damaging agent carbon tetrachloride (CCl4) to induce two different settings of liver regeneration processes in vivo. Liver damage, regeneration and the onset of an inflammatory response will be monitored by measuring the expression of characteristic markers both at the mRNA and protein levels. Local effects in the liver lobules (HSC activation, hepatocyte proliferation, invasion of inflammatory cells, cell death etc) were assessed by immunohistochemical stainings (IHC). Microarray expression profiling and validation by qRT-PCR will as well be performed. At later stages of the project, genetically engineered strains with cell type specific expression of reporter constructs, as described in subproject C1 will be used. Model generation To elucidate molecular mechanisms regulating liver regeneration, it will be important to extend the existing data-based mathematical model of TGF- signalling by including cell cell communication between HSCs and hepatocytes. Furthermore, additional feedback loops besides SnoN (for HSCs especially Smad7 is envisaged) and TGF-/Smad1 signaling as well as Smadindependent signalling is in focus. This will require understanding of the behaviour of HSCs as well as the impact of HSCs to the behaviour of hepatocytes and vice versa, which will be investigated by stimulation with conditioned media and by coculture. Further, it is intended to establish a spatio-temporal model of TGF- mediated signalling in both cell types to uncover dynamic properties such as oscillatory behaviour, time resolved crosstalks with other pathways and link events at the single cell level with hepatocyte regeneration and inflammation at the organ level. The impact of the cell geometry (shape; fibroblastoid versus epithelial polarity) on TGF- mediated signalling will be addressed by advancing inducible vector systems to express fluorescently labelled proteins in hepatocytes/HSCs (more detailes? Which proteins in context of cell geometry? – Ursula?) and by establishing mouse lines expressing fluorescently-labelled variants of signalling components at endogenous levels, as described in subproject C1. Appropriate strategies for spatio-temporal modelling will have to be established (on which basis? phosphorylation status, expression status? Also, is there a concept in which temporal resolution things will be analyzed?). Finally, the goal will be to use these tools to quantitatively link effects at the single cell level with cellular decisions and in conclusion with the regeneration process at the organ level by developing a detailed multi-scale model. This model will facilitate to predict the impact of perturbations and guide the design of experimental setups to understand TGF- signaling during damage, inflammation and regeneration in HSCs and hepatocytes in vitro and at the organ level in vivo. TGF-β regulating negative feedback players are intended to be used to disturb the system and based on arrays, expression patterns (signatures) and quantitative data can be produced. This makes the model more complex, but also closer to the real situation and makes it more trustworthy for “industrial” use. Milestones: M1, SOPs for HSC culture, conditioned media use, HSC-hepatocyte coculture and data generation M2, model of TGF- signaling in quiescent vs activated HSCs a) R-Smad2/3 signaling in quiescent vs activated HSCs (with data mining of published work and based on new data) b) model of R-Smad1 signaling in quiescent vs activated HSCs c) model of negative feedback regulation by Smad7 in quiescent vs activated HSCs M3, model of TGF- signaling in hepatocytes communicating with HSCs a) hepatocytes treated with conditioned media of HSCs (quiescent vs activated) b) hepatocytes cocultured with HSCs M4, model of TGF- signaling in HSCs communicating with hepatocytes a) HSCs treated with conditioned media of hepatocytes b) HSCs cocultured with hepatocytes M5, Model of temporal and spatial distribution of TGF- signaling in mouse liver after partial hepatectomy in comparison to challenge with one single injection of CCl4 a) based on IHC staining with phospho Smad antibodies and quantitative evaluation b) based on gene expression profiling from cultured cells and hepatectomized/CCl4 intoxicated livers M6, Usage of genetically modified animals to generate/refine the model as established in M5 Contribution to economic exploitation (¼ page) There are currently efficient strategies on the market to interfere with TGF- downstream signaling. However, due to the multiplicity of outcomes of this growth factor, which is defined in a strongly contextual manner, no treatment is currently provided for patients. Thus, a spatially (cell type specific) and temporally dissolved model of this signaling pathway in the different phases of liver damage, inflammation and regeneration will be able to temporally define a treatment window and at the same time specify the drugable target of this highly branched signaling network, to perform a TGF- directed therapy. Therefore, the presented application is of high ecomic impact for the pharmaceutical industry. Further, the intended genetically modified animals are of high value for the research community, not only in the liver field, since usage of different promoters for Cre activity may target the genetic modification to any organ of choice. Five major recent subject-related publications of the PI Dooley S, Hamzavi J, Ciuclan L, Godoy P, Ilkavets I, Ehnert S, Ueberham E, Gebhardt R, Kanzler S, Geier A, Breitkopf K, Weng H, Mertens PR: Hepatocyte-specific Smad7 expression attenuates TGF-beta-mediated fibrogenesis and protects against liver damage. Gastroenterology 2008;135:642-659. Weng HL, Ciuclan L, Liu Y, Hamzavi J, Godoy P, Gaitantzi H, Kanzler S, Heuchel R, Ueberham U, Gebhardt R, Breitkopf K, Dooley S: Profibrogenic transforming growth factor-beta/activin receptor-like kinase 5 signaling via connective tissue growth factor expression in hepatocytes. Hepatology 2007;46:1257-1270. Godoy P, Hengstler JG, Ilkavets I, Meyer C, Bachmann A, Muller A, Tuschl G, Mueller SO, Dooley S: Extracellular matrix modulates sensitivity of hepatocytes to fibroblastoid dedifferentiation and transforming growth factor beta-induced apoptosis. Hepatology 2009 Hamzavi J, Ehnert S, Godoy P, Ciuclan L, Weng H, Mertens PR, Heuchel R, Dooley S: Disruption of the Smad7 gene enhances CCL4-dependent liver damage and fibrogenesis in mice. J Cell Mol Med 2008;12:2130-2144. Seyhan H, Hamzavi J, Wiercinska E, Gressner AM, Mertens PR, Kopp J, Horch RE, Breitkopf K, Dooley S: Liver fibrogenesis due to cholestasis is associated with increased Smad7 expression and Smad3 signaling. J Cell Mol Med 2006;10:922-932. Further subject-relevant publications of the PI Stopa M, Benes V, Ansorge W, Gressner AM, Dooley S: Genomic locus and promoter region of rat Smad7, an important antagonist of TGF-beta signaling. Mamm Genome 2000;11:169-176. Stopa M, Anhuf D, Terstegen L, Gatsios P, Gressner AM, Dooley S: Participation of Smad2, smad3, and smad4 in transforming growth factor beta (TGF-beta)-induced activation of Smad7. The TGF-beta response element of the promoter requires functional Smad binding element and ebox sequences for transcriptional regulation. J Biol Chem 2000;275:29308-29317. Dooley S, Delvoux B, Lahme B, Mangasser-Stephan K, Gressner AM: Modulation of transforming growth factor beta response and signaling during transdifferentiation of rat hepatic stellate cells to myofibroblasts. Hepatology 2000;31:1094-1106. Dooley S, Delvoux B, Streckert M, Bonzel L, Stopa M, ten Dijke P, Gressner AM: Transforming growth factor beta signal transduction in hepatic stellate cells via Smad2/3 phosphorylation, a pathway that is abrogated during in vitro progression to myofibroblasts. TGF-beta signal transduction during transdifferentiation of hepatic stellate cells. FEBS Lett 2001;502:4-10. Dooley S, Streckert M, Delvoux B, Gressner AM: Expression of Smads during in vitro transdifferentiation of hepatic stellate cells to myofibroblasts. Biochem Biophys Res Commun 2001;283:554-562. Dooley S, Hamzavi J, Breitkopf K, Wiercinska E, Said HM, Lorenzen J, Ten Dijke P, Gressner AM: Smad7 prevents activation of hepatic stellate cells and liver fibrosis in rats. Gastroenterology 2003;125:178-191.