Our work was originally motivated my collaboration with Drs

Our work was originally motivated my collaboration with Drs
Pasqualini & Arap who have been studying molecular targets that may be used to direct therapies to specific tissues. The long term goal of this kind of work is that if drugs can be targeted to specific tissues in the body, then dosage can be altered to achieve the desired effect while minimizing side effects such as toxicity.
The assumption that different tissues can be targeted presupposes the existence of biochemical differences which can be exploited. The type of difference that we are looking for is a differential concentration of protein binding sites: something that says that if you inject protein A into the body, the only tissue to which that protein can bind is tissue A.
In vivo phage display is a technology used to reveal organ specific vascular ligand-receptor systems in animal models, and recently in patients, and also to validate them as potential therapy targets.
Phage display is a biological process to individually display up to millions of peptides and proteins on the surface of a small bacterial virus -- a phage. The use of phage display in screening for high-affinity ligands and their receptors has been crucial in functional genomics and proteomics.
A bacteriophage library can be constructed to provide the artificial receptors. The library can contain millions of bacteriophage with randomly displayed peptide sequences in the phage outer protein coat which act as binding sites for the agents of interest. This library will be used to 'bio-pan' for phages that bind to a number of toxins and infectious agents and can, thus, provide an endless supply of low cost, reliable, specific, and stable artificial receptors.
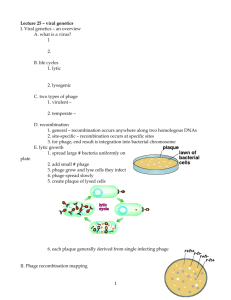
Phage display describes a selection technique in which a library of variants of a peptide or protein is expressed on the outside of a phage virion, while the genetic material encoding each variant resides on the inside (1-3). This creates a physical linkage between each variant protein sequence and the DNA encoding it, which allows rapid partitioning based on binding affinity to a given target molecule (antibodies, enzymes, cell-surface receptors, etc.) by an in vitro selection process called panning
(4). In its simplest form (Figure 1), panning is carried out by incubating a library of phage-displayed peptides with a plate (or bead) coated with the target, washing away the unbound phage, and eluting the specifically bound phage. The eluted phage is then amplified and taken through additional binding/amplification cycles to enrich the pool in favor of binding sequences. After 3-4 rounds, individual clones are characterized by DNA sequencing and ELISA.
Figure 1: Panning with a Ph.D. Phage Display Library
Peptides are composed of amino acids, and the length of a peptide is defined as the number of amino acids in the peptide. For example, a tri-peptide has three amino acids. There are 20 different amino acids, which leads
to 4,200 distinct tri-peptides (not counting mirror images, e.g., ABC is the same as CBA). For quadripeptides, the number of distinct types is approximately
20 times larger. We are looking for differential binding profiles by using specially designed phage probes. A
DNA sequence corresponding to a known peptide sequence is spliced into the phage DNA. When this spliced sequence is transcribed and translated it produces a 9-mer polypeptide of the form $CX_7C$
(here, $C$ = cysteine, $X$ = any amino acid) which is attached to one of the phage's coat proteins.
Conceptually, there are strands extruded from the surface of the phage which terminate in the given polypeptide sequence. The two cysteines at the ends tend to bind to each other, forming a loop. If these loops encounter suitable binding sites, the phages will attach. The experiment proceeds as follows: a large number of phages with diverse
9-mers are injected into the subject and allowed to disperse through the circulatory system. Shortly there after, samples are taken from target tissues of interest. These samples are examined for phage; each phage that is found is cultured and its specific 9-mer sequence determined. Repetitive motifs concentrated in the phage sampled from a particular tissue may indicate the higher availability of a specific type of binding site.
The case study we work with involves 3 stages of mouse filtering. We begin with an unselected cyclic random peptide library CX_7C injected into mice. Six organs were targeted in mice: muscle, bowel uterus, kidney, pancreas, brain. At each stage, peptide-displaying phage are isolated from target organs, amplified, and pooled for the next round of selection. The total number of tri-peptides from a small pool is obtained by randomly sampling from the phage library
harvested from the mouse organs.
the tri-peptide counts from the phage library are expected to increase across the multiple stages of the experiment if they are to bind to specific organs with high affinity. Likewise, the counts for the tri-peptides that do not bind will stay unchanged or even decrease, since only a certain number of peptides may bind to an organ.