Supplementary Material (doc 32K)
advertisement

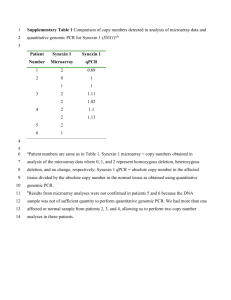
Supplementary Material Supplementary Methods Thermosome primer cocktail evaluation Primer cocktail experiments were conducted to determine if and how JH0175/JH0178 and JH0268/JH0269 could be combined to allow for amplification of a broad spectrum of GC content sequences in a single PCR reaction. A template mixture was created by pooling genomic DNA from Mc. maripaludis, Sulfolobus sp., Tp. acidophilum, Tc. gorgonarius and Hf. volcanii WR341 to a final concentration of 1.0 ng of each genomic DNA per PCR reaction. This template mixture was used with the standard thermosome PCR protocol with molar ratios of 10:0, 7:1, 3:1, 1:1, 1:3, 1:7 and 0:10 JH0175/JH0178:JH0268/JH0269 primers. Independent duplicates of the primary PCR products for each primer ratio mix were generated and each target sequence was quantified in the product by quantitative PCR (qPCR) using standard curves of cloned thermosome sequences. Primary PCR products, as well as the starting template mixture, were diluted 10-1 – 10-5 with water (as necessary to bring the target copy number within range of the standard curve) and added to qPCR reactions containing 400 nM of each species-specific primer (Supplementary table 2) and 1 × iQ SYBR Green Supermix (BioRad, Mississauga, ON), in a final reaction volume of 25 μl. A MyiQ thermocycler was used for all reactions with the following program: 95°C for 3 min, followed by 30 cycles of 95°C for 15 sec, 62°C for 15 sec, 72°C for 15 sec, and a final extension at 72°C for 5 min followed by a dissociation curve. Fluorescent signals were measured every cycle at the end of the annealing step and continuously during the melt curve data collection. Results were analyzed using iQ5 Optical System Software (BioRad). Copy numbers of each target species were quantified, duplicate reactions were averaged and the proportion (mol percent) of each mixture corresponding to each species was Page 1 of 3 calculated before and after thermosome PCR amplification for each of the primer ratio compositions. Supplementary Results Optimization of “universal” to high GC primer ratios for thermosome PCR The optimal ratio of JH0175/JH0178 primers to JH0268/JH0269 primers for use in thermosome PCR was evaluated by determining what cocktail of primers most faithfully amplified a complex community. A mixture of genomic DNA was created to include a broad spectrum of GC content organisms and used as template for thermosome PCR with a range of primer ratios (Supplementary figure 3). The quantity of each constituent of the mixture was determined by qPCR before and after thermosome PCR and expressed as the mol percent of the mixture. For a completely unbiased amplification of the mixture, the ratio of mol percent after thermosome PCR amplification to before PCR amplification should be 1. Supplementary figure 3 illustrates that with only JH0175/JH0178 (10:0 ratio), Mc. maripaludis is preferentially amplified compared to the other species in the mixture (although all species were amplified to some extent). Alternatively, with only JH0268/JH0269 (0:10 ratio), Tp. acidophilum, Tc.gorgonarius and Hf. volcanii were well-represented while Mc. maripaludis and Sulfolobus sp. were not detected after amplification. The 7:1 molar ratio of JH0175/JH0178:JH0268/JH0269 (350 nM/reaction of each JH0175 and JH0178 and 50 nM/reaction of each JH0268 and JH0269) gave the best overall representation, with all targets after amplification being within approximately one log of their starting proportions (Supplementary figure 3). Additional primer ratios of 15:1, 31:1, 63:1, 127:1 and 255:1 were tested but were outperformed by the 7:1 ratio Page 2 of 3 (data not shown). The 7:1 thermosome primer cocktail was tested against the panel of archaeal genomic DNA and was found to generate amplicon from all species tested (Figure 3C). Page 3 of 3