NORWEGIAN UNIVERSITY OF SCIENCE AND TECHNOLOGY
advertisement

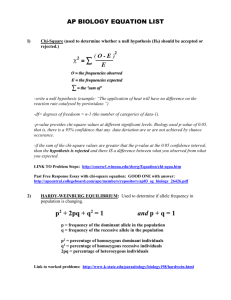
Page 1 of 5 NORWEGIAN UNIVERSITY OF SCIENCE AND TECHNOLOGY Faculty of Natural Sciences and Technology Department of Biology Subject teacher: Jarle Mork (90973351), Hans Stenøien (91897592) EXAMINATION IN: BI 3010 ENGLISH DATE: Points: 7.5 Number of pages: 5 Number of hours: 4 Permitted aids: Calculator HP 30S Grades to be announced on: January 8., 2007 ALL QUESTIONS COUNT AS EQUAL ___________________________________________________________________________ English Question 1. Some very fundamental topics in population genetics theory were formulated already by the pioneers in the field. Among these topics are the Hardy-Weinberg (H-W) theorem, the Wahlund principle, and "Fisher's fundamental theorem on natural selection" (Fisher's theorem). a) Phrase the Hardy-Weinberg theorem in words. b) Phrase the Wahlund principle in words. c) Phrase Fisher's teorem in words. Question 2. Use the information in tables 1 and 2 (Appendix) and answer the following questions: a) What are the allele frequencies for A and B in the samples? b) What are the expected (according to H-W) genotypic numbers in the samples? c) What are the chi-square (Goodness-of-fit test; G-o-f) values for each of the observed genotype distributions? d) How are degrees of freedom (DF) calculated in G-o-f tests? e) What are the significance levels (P) for the observed genotype distributions in the samples (use Appendix Table 2). f) Show how you can use a chi-square RxC contingency table test and Table 2 to decide whether the difference in allelic proportions in samples 1 and 2 is statistically significant, and report your conclusion. Question 3. The so-called "Breeder's equiation" is a simple formula for estimating the expected response (R) in a selection regime, using the heritability (h2) and the selection differential (S) when the population mean and the truncation point (T) is known. (S can also be expressed as I*SDP, where I is the difference (in SDP units) in phenotypic mean values between the population and the selected group). Assume the following scenario: We want to improve the trait <mean length at age=2 years> in a population of farmed fish. In the start population the phenotypic mean value is 40 cm with SDP=8 cm. In one hypothetical experiment (Regime 1), individuals larger than 48 cm are selected for use as brood stock for producing the F1 generation. In a second hypothetical experiment (Regime 2), the largest (by length) 20% of the individuals are used for producing F1. a) What is the expected mean lenght at 2 years in F1 using Regime 1? b) What is the expected mean length at 2 years in F1 using Regime 2? Use Falconer's Table A (Appendix Table 3) as an aid. Question 4. a) Fig. 1 (Appendix Fig. 1) shows clinal variability in Adh allele frequencies in Australian, North American and Eurasian Drosophila melanogaster populations. Each circle represents a population, and solid part of circles represents the proportion of the Fast allele. How will you explain the pattern observed? b) Explain the main features of the “nearly-neutral” theory. c) Explain how phylogenetic trees can be reconstructed by parsimony analysis. d) What is a substitution model? Give examples of two commonly used nucleotide substitution models and explain shortly how they differ. APPENDIX Table 1. Observed genotypic distribution at a locus in samples from two natural population of a diploid organism. Sample 1 AA 40 Sample 2 66 Genotypes AB BB 120 40 110 Table 2. Critical values of chi-square. 24 N 200 200 qA Allele freq. qB Goodness-of-fit test chi-square and DF P Table 3. Falconer's table A Fig 1