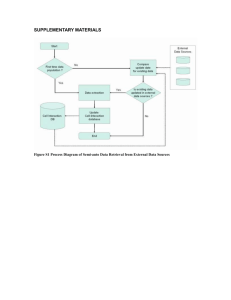
Estimation of Germ-line DNA Copy Number Changes in the Ontario
advertisement

Zogopoulos et al. S- 1 Supplementary Material Supplementary Table 2. Enriched gene ontology categories in CNVRs. Known genes that overlapped with CNVRs were tested for over or under-representation of specific Gene Ontology (GO) gene function annotation terms (The Gene Ontology Consortium 2000) using the BiNGO software (Maere et al. 2005) and updated human GO annotation downloaded on Dec.3.2006. 1351 unique genes were tested against the entire GO reference set, consisting of 16,123 annotated genes. Assessment of significance was conducted using the hypergeometric test and Benjamini & Hochberg False Discovery Rate multiple testing correction. UniProt (Wu et al. 2006) identifiers (IDs) for each known gene were converted to Entrez Gene IDs (Wheeler et al. 2006), using Expasy (Gasteiger et al. 2003) and Ensembl (Clamp et al. 2003). The following GO terms were enriched in CNVRs, while a significant impoverishment of GO categories was not observed. GO-ID Description # of Test Genes in GO Term # of Reference Genes in GO Term Corrected p-value 4984 olfactory receptor activity 64 378 1.08E-04 7608 sensory perception of smell 63 376 1.08E-04 7606 sensory perception of chemical stimulus 64 405 4.99E-04 7600 sensory perception 92 689 2.69E-03 30106 MHC class I receptor activity 9 18 2.69E-03 1584 rhodopsin-like receptor activity 93 726 1.02E-02 7186 G-protein coupled receptor protein signaling pathway 118 1003 3.39E-02 50877 neurophysiological process 109 922 4.58E-02 Zogopoulos et al. Supplementary Material S- 2 Supplementary References: Clamp M, Andrews D, Barker D, Bevan P, Cameron G, Chen Y, Clark L et al. (2003) Ensembl 2002: accommodating comparative genomics. Nucleic Acids Res 31: 38-42 Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31: 3784-3788 Hamosh A, Scott AF, Amberger J, Bocchini C, Valle D, McKusick VA (2002) Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 30: 52-55 Higgins ME, Claremont M, Major JE, Sander C, Lash A E (2006) Cancer Genes: a gene selection resource for cancer genome projects. Nucleic Acids Res 35(Database issue):D721-6. Maere, S., Heymans, K. & Kuiper, M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448-3449. Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM et al. (2006) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 34: D173-D180 Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Mazumder R, O'Donovan C, Redaschi N, Suzek B (2006) The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res 34: D187-D191 Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H et al. (2006) Global variation in copy number in the human genome. Nature 444:444-54 Zogopoulos et al. Supplementary Material S- 3 Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D et al. (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314: 268-74 The Gene Ontology Consortium (2000) Gene ontology: tool for the unification of biology. Nat Genet 25: 25-29.