DOC - Marie Curie Fellows Association
advertisement

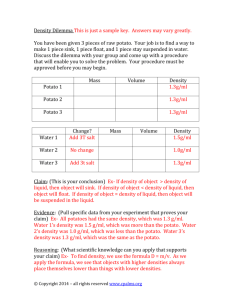
MOLECULAR ANALYSIS OF THE INTERACTION BETWEEN POTATO VIRUS X AND THE RESISTANCE GENE NB IN SOLANUM TUBEROSUM María Rosa Marano, Isabelle Malcuit and David C. Baulcombe The Sainsbury Laboratory, John Innes Centre, Colney Lane, Norwich, NR4 7UH, UK. Contact details of submitting author (name underlined): Tel. 54-341-4370008, Fax. 54-341-4390465 E-mail: marano@ciudad.com.ar Abstract Nb is a single dominant gene in potato that confers hypersensitive resistance to potato virus X group 1 and group 2 (strains ROTH1 and CP2, respectively). Genetic and molecular analysis showed that Nb is located on the upper arm of chromosome V and form part of a cluster of resistance genes encoding specificities to many different pathogens. Part of the strategy for cloning Nb was the construction of a high-resolution genetic map around the Nb locus using amplified fragment length polymorphism (AFLP) technology in conjunction with a bulked segregant approach. We describe the genetical localisation of molecular markers tightly linked to the Nb locus and the development PCR-based markers suitable for isolation of the Nb resistance gene by positional cloning. To characterise the viral elicitor of Nb-mediated resistance we introduced modifications into the genome of the avirulent PVX strain ROTH1 and the virulent PVX strain UK3. We show that the Nb avirulence determinant corresponds to the 25 kDa PVX movement protein and that the isoleucine residue at position 6 in this protein is required for the activation of the Nb response. To study cellular events associated with the Nb response, the 25 kDa proteins of both viral strains were tagged with the green fluorescent protein (GFP). Using laser scanning confocal microscopy we showed that the Nb-mediated response is associated with degradation of subcellular structures. Keywords: Nb resistance gene, Solanum tuberosum, PVX, avirulence gene, map-based cloning María Rosa Marano and Isabelle Malcuit contributed equally to this work. Present address of María Rosa Marano: Instituto de Biología Molecular y Celular (CONICET), Departamento Microbiología, Facultad de Ciencias Bioquímicas y Farmacéuticas, Universidad Nacional de Rosario. Suipacha 531, 2000 Rosario, Argentina. 1 INTRODUCTION Genetic control of resistance to pathogens in plants is often determined by simple gene-for-gene interactions. The resistance response is induced only if the pathogen encodes a strain-specific avirulence (avr) gene and the plant carries the corresponding disease resistance (R) gene [1,2]. In many cases this recognition is manifested as a hypersensitive response (HR) which is associated with a programmed cell death (PCD) at the initial site of infection [3]. Two types of resistance to PVX have been identified in potato: hypersensitive resistance that is controlled by the genes Nb and Nx and extreme resistance (ER) that is conferred by the Rx1 and Rx2 genes. Rx1 and Rx2 are located on chromosome XII and V, respectively [4,5]. Nx maps to chromosome IX [6]. Previous studies have shown that two different features of the PVX coat protein gene are involved in Rx and Nx interactions [7,8]. In this paper, we have focused on the construction of a high resolution genetic map around the Nb locus and on the characterisation of its viral elicitor to study the molecular basis of the Nb-PVX interaction. 2 MATERIALS AND METHODS 2.1 Plant material, virus strains, PVX cDNA clones, expression vectors and resistance assays The tetraploid potato cv Pentland Ivory carrying Nb in the simplex condition (Nb nb nb nb) was self-pollinated to produce an S1 population. PVX strains ROTH1, CP2, CP4 and UK3 were described previously [7,9,10]. ROTH1 and CP2 induce the Nb-mediated HR whereas CP4 and UK3 can infect Nb plants systemically. Plants were tested for resistance to PVX avirulent strains by graft inoculation [11] and/or particle bombardment of detached leaves [10,12]. Plants were considered resistant if they displayed HR (necrotic lesions) on leaves inoculated with ROTH1 or CP2 avirulent strains and if no virus was detected on non-inoculated leaves. Plants were considered susceptible if after challenging with ROTH1 or CP2 the plants became systemically infected with the virus and it could be detected in non-inoculated leaves. ROTH1/UK3 hybrid viral genomes and 25K-GFP fusion constructs have been described previously [10]. 2.2 PCR-based screening of the mapping population and AFLP analysis Genomic DNA for AFLP and PCR-based analyses was isolated according to [5]. Primers, PCR conditions and restriction enzymes used for the markers GP21, SPUD839, TG432 and SPUD237 have been described by [11]. To isolate DNA for the resistant (R) and susceptible (S) bulked segregant pools, equivalent amounts of leaf material from the individuals in each class were pooled before extraction [13]. Template DNA from the R and S pools as well as from each individual recombinant plant was prepared for AFLP as described previously [14,15], using the restriction enzymes PstI and MseI. For selective amplification, 741 primer combinations were analysed: 13 PstI/+2 primers (2 selective nucleotides) and 57 MseI/+3 primers (3 selective nucleotides). AFLP reactions were performed as described by [14]. Bands of interest were cut out of the gel with a scalpel and incubated in 150 µl of TE (10 mM Tris pH 7.5, 1 mM EDTA pH 8.0) overnight at 37˚C. AFLP fragments were recovered by PCR using the same conditions as the initial amplification. PCR products were cloned into the pGEM-T vector (Promega), sequenced and converted to PCR-based markers. The sequences and PCR conditions used for AFLP-derived PCR markers can be obtained on request. resolution map around the Nb locus. This S1 population was screened for recombination events using PCR analysis of DNA samples from individual plants with primers derived from GP21 and SPUD237 markers as described previously [11]. Twenty-six individual plants were identified with recombination events in the interval between markers GP21 and SPUD237. After screening of the recombinant plants the two markers SPUD839 and TG432, initially found to cosegregate with GP21 and SPUD237 respectively [11], could be separated from GP21 and SPUD237 by 3 and 2 recombination events (Fig. 1A). A Plant Number: 1 3 1 3 3 8 3 Distances R S A B C (cM) D E F G 3 1 Markers H I GP21-11 AFLPs 0.23 (3) SPUD839 2 AFLPs 0.85 (11) GM339 0.30 (4) Nb 0.46 (6) 0.16 (2) B M TG432-GM6372 AFLPs SPUD237 26 AFLPs GM339 330 bp 2.3 Bombardment of detached potato leaves and confocal microscopy experiments The methods employed for transient expression using particle bombardment have been described previously [10,12]. Imaging of GFP fluorescence was performed with a LEICA TCS-NT confocal laser scanning microscope equipped with a 20X or 40X dry objective. Laser illuminations at 488 and 568 nm (Argon ion laser) were recorded through a 530 nm or 600 to 630 nm bandpass filter. 3 RESULTS 3.1 Selection of recombinants in the GP21SPUD237 interval Nb was previously mapped on chromosome V in a 3.3 centimorgan (cM) interval delimited by markers GP21/SPUD839 and TG432/SPUD237 [11]. We extended the original mapping population to a total of 1300 S1 progeny to allow the construction of a high 220 bp GM637 Figure 1: High-resolution genetic map of the Nb locus. A. Map position of the Nb locus based on the screening of 741 AFLP primer combinations for AFLP markers linked to the Nb locus and 1300 segregant plants for recombination events in the interval GP21SPUD237. The genetic distance (in cM) corresponds to the percent of recombination between molecular markers or molecular markers and Nb in a population of 1300 plants. B. Shows the analysis of individuals with recombination events in the GP21-SPUD237 interval using the GM339 and GM637 markers. The resistant pool and susceptible pools are indicated as R and S respectively. Arrows show the sizes of the markers Each recombinant plant was then tested for resistance to PVX avirulent strains CP2 or ROTH1 and Nb was mapped between SPUD839 and TG432 at 1.15 cM and 0.46 cM, respectively (Fig. 1A). 3.2 High-resolution genetic map around the Nb locus To identify DNA markers in the region delimited by the markers SPUD839 and TG432, containing Nb, AFLP technology [15] was employed in conjunction with a bulked segregant approach [13]. A total of 741 random selected PstI/MseI primer combinations were analysed and 69 of those revealed polymorphisms between the R and S pools. To map these new polymorphic markers relative to SPUD839 and TG432, the AFLP analysis was repeated using the recombinants detected in the GP21 and SPUD237 interval. Of these, 11 AFLP markers cosegregated with GP21, 2 AFLP markers (GM715 and GM1350, Fig. 1A) co-segregated with SPUD839, 1 AFLP marker (GM339, Fig. 1A) was mapped between SPUD839 and Nb, 3 AFLP markers (GM637, GM1216 and GM1219, Fig. 1A) co-segregated with TG432, 26 AFLP markers co-segregated with SPUD237 and the remaining 26 markers were mapped outside the GP21SPUD237 interval (Fig. 1A). The result of this analysis places Nb in an interval of 0.76 cM, flanked by the marker GM339 and the cosegregating markers TG432, GM637, GM1216 and GM1219 (Fig. 1A). To develop a fast screening method, the AFLP markers closest to Nb and bigger in length, GM339 and GM637, were converted to PCR markers. GM339-derived primers amplified a 330 bp fragment and GM637-derived primers a 220 bp fragment (Fig. 1B). The GM339 and GM637 primers amplified only the resistant allele and were used to analyse the 26 plants with recombination events between GP21 and SPUD237. The results obtained from this analysis were in accordance with the map presented in Fig. 1. virulent strain UK3, or with the wild-type virulent control pUK3 (Fig. 2). A B ROTH1 Replicase . Ile6 25K 8K CP 12K UK3 Ser6 REP31 REP13 FA313 FA131 UK3-Ile6 Ile6 3.3 Mapping of the Nb avirulence determinant in the PVX genome To identify the PVX avirulence determinant that is involved in the activation of Nb-mediated HR in potato, two viral strains were selected, ROTH1 [10] and UK3 [7]. To map the Nb avirulence determinant in the PVX genome, a set of hybrid viruses was constructed by exchanging cDNA sequences between the avirulent strains ROTH1 and the Nb resistance-breaking strain UK3 (Fig. 2A). The interaction between Nb and the ROTH1/UK3 viral hybrids was analysed by particle bombardment of these chimeric genomes on potato plants. Bombardment of detached resistant (Nb) potato leaves with viral cDNA hybrids carrying the 25K gene from ROTH1 (REP31 and FA313) or the wild-type avirulent control pROTH1 led to the formation of necrotic lesions 3 days after bombardment, indicative of an interaction between Nb and the corresponding PVX avirulence determinant (Fig. 2). In contrast, there were no hypersensitive lesions with REP13 and FA131, containing the 25K gene from the Figure 2: Analysis of ROTH1/UK3 hybrids and UK3-Ile6 mutant by particle bombardment of detached Nb potato leaves. A. Schematical representation of the wild type and modified PVX genomes. REP31, REP13, FA313 and FA131 were generated by exchanging cDNA fragments between the PVX avirulent strain ROTH1 and the Nb resistance-breaking strain UK3 [10]. UK3-Ile6 is a viral mutant derived from UK3 in which the serine residue at position 6 in the 25 kDa protein is replaced by isoleucine [10]. B. Responses induced on Nb leaves three days after bombardment with the hybrid and mutant cDNA constructs. HR is manifested by the appearance of necrotic lesions (black spots) at the site of impact. All of these PVX chimeras induced the formation of symptoms in susceptible (nb) potato plants and accumulated to the same level as the wild-type strains, as measured by ELISA detection of the viral coat protein [10]. Therefore, induction of the Nb-mediated resistance in potato is dependent upon either the PVX 25 kDa movement protein or its mRNA. Comparative sequence analysis of the 25 kDa proteins of ROTH1, UK3, CP2, and CP4 identified 32 amino acid substitutions in total [16]. However, there is only one amino acid difference when the 25 kDa protein sequences of ROTH1 and CP2 were compared to that of UK3 and CP4 respectively [10,16]. The isoleucine at position 6 (Ile6) in the 25 kDa protein of ROTH1 and CP2 is replaced by a serine (Ser6) in UK3 or a threonine (Thr6) in CP4 [10]. This conservation of Ile6 in the two avirulent strains indicates that it is probably essential for activation of the Nb response. To test this hypothesis, a mutant transcription clone was generated, resulting in a single substitution in the UK3 25 kDa protein sequence at position 6 (Ser6Ile6: UK3-Ile6 mutant) [10]. When this construct was tested on plants, UK3-Ile6 induced Nbmediated necrosis (Fig. 2), whereas UK3 produced systemic chlorotic symptoms in graft-inoculated Nb plants (Fig. 2). These results indicate that the presence of Ile 6 in the 25 kDa protein is required for activation of the Nb response. of subcellular structures specifically associated with Nbmediated HR. A 25K1 GFP p25K3GFP 25K3 GFP Nb 3.4 Dynamics of the subcellular distribution of 25KGFP fusion proteins in relation to the Nbmediated response To investigate the cellular processes associated with pathogen-induced HR, GFP was used as a marker [17] to follow the localisation of the 25 kDa protein in potato cells and monitor cellular changes specifically associated with the activation of Nb-mediated HR. This gene was fused to the 3’ end of the 25K gene of ROTH1 (p25K1GFP) and UK3 (p25K3GFP), (Fig. 3). GFP fluorescence was detected 6 hours after bombardment in epidermal cells for all the constructs tested (Fig. 3). Free GFP expressed from the CaMV 35S promoter (pGFP construct) accumulated in the nucleus and was found in the cytoplasm, as described previously [17] (Fig. 3C). In contrast, GFP fluorescence in potato cells expressing the 25K1GFP and 25K3GFP fusion proteins was initially localised in inclusions in the cytoplasm of Nb and nb cells and also seemed to be present in or associated with the nucleus (Fig. 3A,B). High resolution imaging of 25KGFP inclusions revealed a network of Y-shaped tubular structures (Fig. 3A). Eventually, starting between 28 and 34 hours after bombardment, these inclusion bodies fragmented into progressively smaller pieces in Nb potato cells bombarded with the avirulent construct p25K1GFP (Fig. 3A). After 48 hours, there was no remaining GFP fluorescence in Nb cells bombarded with p25K1GFP, indicating that the cell death process was complete (data not shown). In control experiments involving bombardment of nb plants with p25K1GFP and Nb plants with p25K3GFP, the green fluorescence was still visible after 48 hours and the large inclusions remained intact (Fig. 3B). These data suggest that the changes in GFP fluorescence reflect degradation p25K1GFP nb B Nb C pGFP GFP Nb Figure 3: Real time imaging of the subcellular distribution of 25KGFP fusion proteins in Nb and nb potato cells. GFP fluorescence was detected by laser scanning confocal microscopy. Each picture shows the distribution of GFP fluorescence in different epidermal cells at different times after bombardment of detached Nb and nb potato leaves with p25K1GFP (A), p25K3GFP (B) and pGFP (C). The CaMV 35S promoter and terminator sequences are indicated by purple and brown boxes respectively. h: hours post-bombardment. The position of the nucleus (n) is indicated by an arrow. A large inclusion body (i) associated with the nucleus is composed of a network of Y-shaped tubular structures. 5 DISCUSSION In this report we have identified new molecular markers tightly linked to the Nb locus and characterised the PVX elicitor of the Nb-mediated response. In the high resolution genetical map described above, we positioned the Nb locus in an interval of approximately 0.76 cM between the AFLP markers GM339 and GM637 (Fig. 2A). Given that the average recombination frequency in potato is about 1000 kb.cM-1 [18], the distance between the closest flanking markers should correspond to approximately 760 kb. However, previous studies in potato have shown that the relationship between genetical and physical distances can vary considerably. For instance, in the case of potato cultivar Cara, which was used to isolate the Rx1 gene, recombination frequencies were found to vary from 180 kb.cM -1 to 2677 kb.cM-1, estimated from the number of recombination events in individual BAC clones [19]. Therefore, the physical distance in the genetical interval between markers GM339 and GM637 cannot be estimated accurately at this stage. However, the small number of AFLP markers identified near the GP21 and SPUD237 interval and their resistant-allele specificity may indicate that the physical distance in this genetic interval is not very large. Future experiments towards the isolation of Nb will focus on the screening of a BAC library from potato cultivar Pentland Ivory with the new tightly linked markers. As part of our strategy to analyse the Nb/PVX interaction we have also shown that the PVX 25 kDa protein is the elicitor of Nb-mediated HR. In compatible interactions the role of this protein is to facilitate cell-to-cell movement of the virus, probably by interacting with plasmodesmata [20,21]. At this stage we do not know whether these two different functions are inter-related. It is also possible that Nb, like other resistance genes, is part of a surveillance system for detection of foreign molecules in plant cells [22]. Analysis of both resistance-inducing (ROTH1) and resistance-breaking (UK3) strains showed that the elicitor function involves the isoleucine at position 6 (Ile6) in the 25 kDa protein. We used 25KGFP fusions to study subcellular distribution of the PVX 25 kDa protein in living potato cells, and to monitor its fate and other cellular events associated with Nb-mediated cell death. The HR-eliciting property of the 25 kDa protein allowed the visualisation by laser scanning confocal microscopy of a cellular process specifically associated with the Nbmediated response. The large GFP-labelled perinuclear bodies in the bombarded potato cells expressing the 25KGFP fusion may represent the lamellar inclusions detected previously by immunocytochemistry and in ultrastructural studies [23]. These inclusion bodies are intimately associated with the endoplasmic reticulum and with actin microfilaments [24]. As the Nb-elicited cells died, these perinuclear structures fragmented and disappeared. In animal cells, programmed cell death was associated with cleavage of microfilament proteins by caspases [25]. Previous studies in plants have implicated caspase-like enzymes in disease resistance [26]. Therefore, it is possible that the fragmentation of the 25K1GFP inclusion bodies during the Nb-mediated response is due to the action of a caspase-like protease and/or the disruption of actin microfilaments. The cloning and characterisation of the Nb gene might reveal new domains involved in the recognition of pathogen elicitors and the explanation about the difference between HR provided by Nb and ER conferred by the Rx gene to PVX, that are elicited by two different virus-encoded proteins, respectively the 25 kDa protein and the coat protein [10,27]. In addition, Nb is particularly important because of its genetic linkage to other loci conferring resistance against fungus, nematodes and other viruses [11,28-30]. Consequently, the isolation of Nb and the physical analysis of this cluster of R genes will provide a good experimental system to study evolution of R genes in potato and the mechanisms by which plants generate new recognition specificities. 6 ACKNOWLEDGEMENTS We thank Gianinna Brigneti for critical reading of the manuscript. M.R.M. was supported by a Marie Curie Research Training Grant no: ERBFMBICT 961303 and is a MCFA member, MN3207. Sainsbury Laboratory is supported by the Gatsby Charitable Foundation. 7 REFERENCES [1]H. H. Flor (1971). "Current status of the gene-for-gene concept". Annu.Rev.Phytopathol. 9, 275-298. [2]N. T. Keen (1990). "Gene-for-gene complementarity in plantpathogen interactions". Annu.Rev.Genet. 24, 447-463. [3]J.-B. Morel, and J. L. Dang (1997). "The hypersensitive response and the induction of cell death in plants". Cell Death and Differentiation 4, 671-683. [4]E. Ritter, T. Debener, A. Barone, F. Salamini, and C. Gebhardt (1991). "RFLP mapping on potato chromosomes of two genes controlling extreme resistance to potato virus X (PVX)". Mol.Gen.Genet. 227, 81-85. [5]A. Bendahmane, K. V. Kanyuka, and D. C. Baulcombe (1997). "High resolution and physical mapping of the Rx gene for extreme resistance to potato virus X in tetraploid potato". Theor.Appl.Genet 95, 153-162. [6]T. J. Tommiska, J. H. Hamalainen, K. N. Watanabe, and J. P. T. Valkonen (1998). "Mapping of the gene Nx(phu) that controls hypersensitive resistance to potato virus X in Solanum phureja lvP35". Theor.Appl.Genet 96, 840-843. [7]T. Kavanagh, M. Goulden, S. Santa Cruz, S. Chapman, I. Barker, and D. C. Baulcombe (1992)."Molecular analysis of a resistancebreaking strain of potato virus X". Virology 189, 609-617. [8]S. Santa Cruz, and D. C. Baulcombe (1993). "Molecular analysis of potato virus X isolates in relation to the potato hypersensitivity gene Nx". Mol.Plant-Microbe Interact. 6, 707-714. [9]B. E. Orman, R. M. Celnik, A. M. Mandel, H. N. Torres, and A. N. Mentaberry (1990). "Complete cDNA sequence of a South American isolate of potato virus X". Virus Res. 16, 293-306. [10]I. Malcuit, M. R. Marano, T. A. Kavanagh, W. de Jong, A. Forsyth, and D. C. Baulcombe (1999). "The 25-kDa movement protein of PVX elicits Nb-mediated hypersensitive cell death in potato". Mol.PlantMicrobe Interact. 12, 536-543. [11]W. de Jong, A. Forsyth, D. Leister, C. Gebhardt, and D. C. Baulcombe (1997). "A potato hypersensitive resistance gene against potato virus X maps to a resistance gene cluster on chromosome V". Theor.Appl.Genet 95, 246-252. [12]M. R. Marano, and D. C. Baulcombe (1998). "Pathogen-derived resistance targeted against the negative-strand RNA of tobacco mosaic virus: RNA strand-specific gene silencing?" Plant J. 13, 537-546. [13]R. W. Michelmore, I. Paran, and R. V. Kesseli (1991). "Identificatiion of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations". Proc. Natl. Acad. Sci. USA 88, 9828-9832. [14]C. M. Thomas, P. Vos, M. Zabeau, D. A. Jones, K. A. Norcott, B. P. Chadwick, and J. D. G. Jones (1995). "Identification of amplified restriction fragment polymorphism (AFLP) markers tightly linked to the tomato Cf-9 gene for resistance to Cladosporium fulvum". Plant J. 8, 785-794. [15]P. Vos, R. Hogers, M. Bleeker, M. Reijans, T. Van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, and M. Zabeau (1995). "AFLP - A new technique for DNA - fingerprinting". Nucleic Acids Res. 23, 4407-4414. [16]I. Malcuit, W. de Jong, D.C. Baulcombe, D.C. Shields, and T.A. Kavanagh (2000) "Acquisition of multiple virulence/avirulence determinants by Potato Virus X (PVX) has occurred through convergent evolution rather than through recombination". Virus genes. 20, 165172. [17]J. Haseloff, K. R. Siemering, D. C. Prasher, and S. Hodge (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122-2127. [18]S. D. Tanksley, M. W. Ganal, J. P. Prince, M. C. Devicente, M. W. Bonierbale, P. Broun, T. M. Fulton, J. J. Giovannoni, S. Grandillo, G. B. Martin, R. Messeguer, J. C. Miller, L. Miller, A.H. Paterson, O. Pineda, M. S. Roder, R. A. Wing, W. Wu, and N. D. Young (1992). "High-density molecular linkage maps of the tomato and potato genomes". Genetics 132, 1141-1160. [19]K. Kanyuka, A. Bendahmane, N. A. M. Jeroen, R. van der Voort, E. A. G. van der Vossen, and D. C. Baulcombe (1999). "Mapping of intralocus duplications and introgressed DNA: aids to map-based cloning of genes from complex genomes illustrated by analysis of the Rx locus in tetraploid potato". Theor.Appl.Genet 98, 679-689. [20]S. M. Angell, C. Davies, and D. C. Baulcombe (1996). "Cell-to-cell movement of potato virus X is associated with a change in the size exclusion limit of plasmodesmata in trichome cells of Nicotiana clevelandii". Virology 215, 197-201. [21]S. Y. Morozov, O. N. Ferdorkin, G. Juttner, J. Schiemann, D. C. Baulcombe, and J. G. Atabekov (1997). "Complementation of a potato virus X mutant mediated by bombardment of plant tissues with cloned viral movement protein genes". J.Gen.Virol. 78, 2077-2083. [22]J. G. M. de Wit (1997). "Pathogen avirulence and plant resistance: a key role for recognition". Trend in Plant Sci. 2, 452-458. [23]C. Davies, G. J. Hills, and D. C. Baulcombe (1993). "Subcellular localisation of the 25kDa protein encoded by the triple gene block of potato virus X". Virology 197, 166-175. [24]I. Malcuit, M.R. Marano, and D.C. Baulcombe (1999). "GFP as a tool to study cellular changes associated with the Nb-mediated hypersensitive response". 9th International Congress on Molecular PlantMicrobe Interactions, Amsterdam, the Netherlands. [25]C. Brancolini, M. Benedetti, and C. Schneider (1995). "Microfilament reorganisation during apoptosis: the role of Gas2, a possible substrate for ICE-like proteases". EMBO J. 14, 5179-5190. [26]O. del Pozo, and E. Lam (1998). "Caspases and programmed cell death in the hypersensitive response of plants to pathogens". Curr. Biol. 8, 1129-1132. [27]A. Bendahmane, B. A. Köhm, C. Dedi, and D. C. Baulcombe (1995). "The coat protein of potato virus X is a strain-specific elicitor of Rx1-mediated virus resistance in potato". Plant J. 8, 933-941. [28]C. Leonards-Schippers, W. Gieffers, F. Salamini, and C. Gebhardt, (1992). "The R1 gene conferring race-specific resistance to Phytophthora infestans in potato is located on potato chromosome V". Mol.Gen.Genet. 233, 278-283. [29]C. M. Kreike, J. R. A. De Koning, J. H. Vinke, J. W. Van Ooijen, and W. J. Stiekema (1994). "Quantitatively inherited resistance to Globodera pallida is dominated by one major locus in Solanum spegazzinii". Theor. Appl.Genet 88, 764-769. [30]J. Rouppe van der Voort, P. Wolters, R. Folkertsma, R. Hutten, P. van Zandvoort, H. Vinke, K. Kanyuka, A. Bendahmane, E. Jacobsen, R. Janssen, and J. Bakker (1997). "Mapping of the cyst nematode resistance locus Gpa2 in potato using a strategy based on comigrating AFLP markers". Theor.Appl.Genet 95, 874-880.